|
Endodontic Topics 2003, 6, 3–28 |
Copyright © Blackwell
Munksgaard |
Life as an endodontic pathogen
Ecological differences between the untreated and root-filled root canals
GÖRAN SUNDQVIST & DAVID FIGDOR
This review describes the type of microbial flora in the untreated root canal and the root-filled canal with persistent infection. Recent contributions of molecular methods of microbial identification are outlined along with a discussion of advantages and limitations of these methods. Ecological and environmental factors are the prime reasons for differences in the microbial flora in these distinct habitats. Shared phenotypic traits and an ability to respond to the modified environment select for the species that establish a persistent root canal infection.
Introduction
Life is not easy for an endodontic pathogen. Microbes seeking to establish in the root canal must leave the nutritionally rich and diverse environment of the oral cavity, breach enamel, invade dentine, overwhelm the immune response of the pulp and settle in the remaining necrotic tissue within the root canal. During that time they have to compete in a limited space with other microbes for the available nutrition. It is no accident that microbes berth in a particular environment – there are ecological advantages for them to establish and flourish if conditions are favorable. Through genetic exchange and mutation, microbes have developed specialized systems that facilitate their ability to find, compete and survive in these very specific environments.
Bacteria are everywhere, but the environment selects
In the oral cavity, there are an estimated 1010 bacteria (1) consisting of more than 500 different kinds of microorganisms (2, 3) and all seek a niche and nutrition. One of the primary functions of tooth enamel is to exclude these microorganisms from the underlying dentine–pulp complex. As long as the enamel and cementum layers are intact, the pulp and root canal are protected from invasion, but loss of these structures by caries, cracks or trauma opens an avenue for penetration of bacteria through the dentinal tubules. All bacteria within the oral cavity share the same opportunities for invading the root canal space; however only a restricted group of species have been identified in infected root canals (4–7). The reason for the disproportionate ratio between potential and actual number of species is that the root canal is a unique environment where biological selection drives the type and course of infection. An anaerobic milieu, interactions between microbial factors and the availability of nutrition are principal elements that define the composition of the microbial flora.
In the initial phase of a root canal infection, the number of species is usually low. If the way of invasion is via caries, the bacteria in the front of the carious process are the first to reach the pulp. In cases where there is no apparent communication with the oral cavity and the bacteria penetrate through dentinal tubules, as in trauma cases without pulp exposure, there is no clear pattern of primary bacterial invaders (4, 5). The number of bacterial species in an infected root canal may vary from one to more than 12, and the number of bacterial cells varies from <102 to>108 per sample. A correlation seems to exist between the size of the periapical lesion and the number of bacterial species and cells in the root canal. Teeth with long-standing infections and large lesions usually harbor more bacterial species and have a higher density of bacteria in their root canals than teeth with small lesions.
The oral and root canal flora
Most of the resident oral microbial flora is consistent with dental health. The predominant microbial diseases of the oral cavity, caries and periodontal disease, develop at sites where a microbial biofilm, plaque, is already established and disease occurs with a change in the environmental conditions, the type and mix of microbial flora. Thus, changes at the tooth surface with a buildup of acidogenic or aciduric bacteria result in demineralization at the tooth surface, leading to caries. An increase in proteolytic bacteria at the gingival crevice is one of a sequence of factors leading to the development of periodontal disease (8). Of the major dental diseases, infection of the root canal is unique for the oral cavity since infection establishes where no microorganisms have previously been present.
The root canal as a unique site of infection
In 1894, WD Miller published his findings on the bacteriological investigation of pulps (9). He observed many different microorganisms in the infected pulp space and realized that some were uncultivable when compared with the full range observed by microscopy, and that the flora was different in the coronal, middle and apical parts of the canal system. Due to limitations of his sampling and cultivation technique, Miller was unable to verify this observation and it was not until 1982 that this could be shown by culturing (10). Differences in availability of nutrients and oxygen tension in the apical region compared with the main root canal are important reasons for the dominance of slow growing, obligately anaerobic bacteria in the apical region.
Studies on the dynamics of root canal infections have shown that the relative proportions of anaerobic microorganisms and bacterial cells increase with time and that the facultatively anaerobic bacteria are outnumbered when the canals have been infected for 3 months or more (10). When a combination of bacterial strains originally isolated from an infected root canal were inoculated in equal quantities into further canals in experimental infections, the original proportion of bacterial strains was reproduced and anaerobic bacteria dominated again (11). This illustrates that interactive mechanisms operate amongst these microorganisms, a concept further supported by the finding that when Prevotella oralis (formerly Bacteroides oralis) was inoculated on its own it was unable to survive, whereas when inoculated with other bacteria it survived and dominated the established flora (11). These experiments have shown that the endodontic milieu is a selective habitat that supports the development of specific proportions of the anaerobic microflora. Oxygen and oxygen products play an important role as ecological determinants in the development of specific proportions of the root canal microflora (12–14). The consumption of oxygen and production of carbon dioxide and hydrogen along with the development of a low reduction–oxidation potential by the early colonizers favor the growth of anaerobic bacteria.
Nutrition as an ecological driver
The type and availability of nutrients is important in establishing microbial growth. Nutrients may be derived from the oral cavity, degenerating connective tissue (13), dentinal tubule contents, or a serum-like fluid from periapical tissue (15). These factors in the root canal environment permit the growth of anaerobic bacteria capable of fermenting amino acids and peptides, whereas bacteria that primarily obtain energy by fermenting carbohydrates may be restricted by lack of available nutrients. This is the likely reason why the flora is dominated by facultatively anaerobic bacteria, such as streptococci, in the coronal section of root canals exposed to the oral cavity, and anaerobic bacteria dominate in the apical section (9, 10).
The succession of strict over facultative anaerobes with time (10, 11) is most likely due to changes in available nutrition, as well as a decrease in oxygen availability. Facultatively anaerobic bacteria grow well in anaerobiosis; however, their prime energy source is carbohydrates. A decrease in availability of carbohydrates in the root canal occurs when there is no direct communication with the oral cavity, which severely limits growth opportunities for facultative anaerobes.
The experiments of ter Steeg and van der Hoeven (16) offer important clues about the likely dynamics of the root canal flora. Using serum as a substrate, they studied the succession of subgingival plaque organisms during enrichment growth. Three phases could be distinguished during growth. Initially, rapidly growing saccharolytic bacteria consumed the low levels of carbohydrates in serum, leading to lactic and formic acid production. In a second phase, proteins were hydrolyzed, some amino acid fermentation took place, and there was digestion of remaining carbohydrates. Carbohydrates were split off from the serum glycoproteins. Growth during this phase was dominated by Prevotella intermedia, Veillonella parvula, Fusobacterium nucleatum and Eubacterium species. In a final phase, there was progressive protein degradation. The predominant species during this phase were Peptostreptococcus micros, F. nucleatum, and eubacteria. The dominance of P. micros in cultures originating from subgingival microbiota, when grown in serum, has also been shown in another study (17). The ecological niche of P. micros may be related to its wide range of peptidase activities, making amino acids and peptides available from serum glycoproteins (16). These amino acids can be used by P. micros, but also by other bacteria that have little or no proteolytic activity in serum.
The black-pigmented anaerobic rods, Prevotella intermedia/nigrescens, Porphyromonas gingivalis and Porphyromonas endodontalis, are proficient in degrading serum proteins and make peptides and amino acids available for fermentation (18). The degradation of native proteins by Prevotella and Porphyromonas species enables the growth of bacteria that depend on the availability of peptides, such as eubacteria, fusobacteria and peptostreptococci, which produce peptidases but cannot hydrolyze intact proteins (19, 20). This is also of importance for the capacity of root canal bacteria to induce periapical abscesses. Combinations of P. micros with P. intermedia or P. endodontalis have been implicated in the induction of periapical abscesses (21). Abscesses harboring a microflora that rapidly degrade serum proteins have been shown to be nearly three times larger than abscesses with a microflora that lack the capacity for breakdown of serum proteins (22).
Growth of mixed bacterial populations may depend on a food chain in which the metabolism of one species supplies essential nutrients for the growth of other members of the population (19, 23–26). Black-pigmented anaerobic rods (Prevotella and Porphyromonas species) are examples of bacteria that have very specific nutritional requirements. They are dependent on vitamin K and hemin for growth. Vitamin K can be produced by other bacteria (27). Hemin becomes available when hemoglobin is broken down, but some bacteria may also produce hemin. Another example is Campylobacter rectus which can stimulate the growth of Porphyromonas species by producing a growth factor related to hemin (26). C. rectus itself derives a source of energy from the co-inhabiting microbial species. It is strictly dependent on a respiratory mechanism in which only formate and hydrogen can serve as electron donors and fumarate, nitrate, or oxygen as electron acceptors. This makes this organism dependent on bacteria producing formate or hydrogen. A wide range of nutritional interactions is recognized among oral bacteria and these may also influence the associations between bacteria in the root canal (28–30).
Because the nutritional supply governs the dynamics of the microbial flora, it means that the bacteria present in the root canal will depend on the stage of the infection. Initially, there may be no clear associations between bacteria, but strong positive associations develop among a restricted group of the oral flora due to the type of nutrients in the environment (28, 31–33). Thus, F. nucleatum is associated with P. micros, P. endodontalis and C. rectus (28). Strong positive associations exist between P. intermedia and P. micros (28, 33) and P. anaerobius (28). There is also a positive association between P. intermedia, and P. micros, P. anaerobius and the eubacteria (28). In general, species of Eubacteria, Prevotella and Peptostreptococcus are positively associated with one another in endodontic samples (28, 31, 33). Properties that these bacteria have in common are that they ferment peptides and amino acids and are anaerobic (16), which indicates that the main source of nutrition in root canals are tissue remnants and a serum-like substrate.
Most of the species isolated from infected root canals have also been identified in the periodontal pocket (2). Although the root canal flora is not as complex as that of the periodontal pocket, they are similar in that both contain a special and limited assortment of the oral flora. It is worth noting, however, that there are some differences. When the microbial flora from root canals and periodontal pockets of non-vital teeth with periodontitis were compared (34), the ratio between anaerobic and facultatively anaerobic bacteria was approximately 100 times higher in the root canal. Anaerobic streptococci were frequent in the root canals and rarely found in the periodontal pockets. In contrast, facultative streptococci outnumbered anaerobic streptococci in the periodontal pockets. Bacteria found in root canals, but rarely in periodontal pockets were Peptostreptococcus species, Pseudoramibacter alactolyticus (formerly Eubacterium alactolyticum), P. intermedia and V. parvula. Bacteria commonly detected in periodontal pockets, but not found or found in low frequency in root canals were Prevotella melaninogenica, Prevotella corrodens and P. gingivalis.
Methods for isolation and cultivation
In order to identify microorganisms and study their characteristics and pathogenic potential, it is essential that accurate methods be used for sampling and cultivation. Microbial cells must survive the accumulated stress of sampling, dispersion, oxygen exposure, and lack of suitable nutrients in the culture media.
Anaerobic bacteriological techniques are indispensable for sampling and cultivation of endodontic bacteria. When the Virginia Polytechnic Institute (VPI) developed and simplified (35) the methods initially developed by Hungate (36), it resulted in a changed picture of the periodontal pocket flora and root canal flora. Application of these techniques to endodontic samples revealed that obligate anaerobes dominated infected root canals, comprising as much as 90% of the flora (4, 6) and confirmed the unequivocal importance of bacteria for the development of apical periodontitis in human teeth (4).
The safest way to protect anaerobic bacteria is to avoid exposure to oxygen during the various phases of the sampling and laboratory work. Oxygen is toxic because of the formation of hydrogen peroxide, superoxide radicals and hydroxyl radicals (37). The VPI method was based on achieving a low reduction–oxidation potential by gassing media with oxygen-free gas, thus affording protection from oxygenation during sterilization and subsequent handling. Another way to protect bacteria from oxygen is to use an anaerobic glove box with an atmosphere containing a mixture of nitrogen, carbon dioxide and hydrogen. Under certain conditions, anaerobic bacteria may tolerate a short exposure to oxygen (14). Media that contain hemolyzed blood have a protective effect (14). The protective effect of blood depends on the presence of the enzyme catalase, which splits hydrogen peroxide in the medium.
Because the number of bacterial cells in samples from infected root canals is so high, samples must be diluted and cultivated on solid media to allow diverse species to grow and be identified as single colonies. The dilution process is the most critical step with regard to oxygen exposure of the sample. Ideally, an anaerobic box and pre-reduced dilution solutions are used, but it can also be performed on the bench by gassing jars with oxygenfree gas. Bacteria isolated from root canals have different thresholds of sensitivity to oxygen. The most sensitive are P. anaerobius, P. endodontalis and Fusobacteria species. The rate of isolation of these species is a good indicator of how careful the measures have been to protect the sample from oxygen exposure during handling (14).
There is a correlation between the size of the periapical lesion and the number of bacterial cells and species in the root canal (4, 28). Samples taken from teeth with large, long-standing lesions harbor more bacteria (up to 108 cfu) than smaller lesions. Different colony types are most easily recognized on plates with 100 or less colonies, but additional colony types are often found on plates with up to 300 colonies. Therefore, it is reasonable to calculate that the detection level for different species is 0.3–1%. For some species, however, this level maybe lowered to 0.01% by using selective media (38).
The number of bacterial cells is dramatically reduced during root canal instrumentation and irrigation (39, 40), so for detection of bacterial cells in these samples it is necessary to inoculate solid media with undiluted samples and to use enrichment growth media, since some samples can contain less than 102 cells (41, 42). Similarly, in root canals of teeth with post-treatment disease, the number of surviving cells can be low, and all precautions must be taken to detect the bacteria. It is advisable to use as small amount as possible of sampling solution to avoid dilution of the sample, then inoculate an undiluted sample on some plates and use enrichment growth (43–45). The detection level for bacteria in teeth undergoing re-treatment depends on what bacterial species are present and on how the sample is treated, but 10–102 cells might be expected in the sample.
Cultivation discloses the broadest spectrum of the endodontic flora. When all the critical steps are well performed, cultivation allows detection of small numbers of cells in a sample. However, in samples with a high cell density the process of serial dilution means that individual species may go undetected if a species is low in number. The methods for detection of contamination, for example leaking rubber dam, or growth of bacteria under a restoration have all been based on cultivation of control samples taken after isolation, access preparation and treatment (43, 46–48).
What is an endodontic pathogen?
Over the last century, the concept of what constitutes a microbial pathogen has changed with an increasing appreciation of the complexity of the host–pathogen interaction. Recently, a revised definition has been proposed by which a pathogen is ‘a microbe capable of causing host damage’ where ‘host damage can result from either direct microbial action or the host immune response’ (49).
An endodontic pathogen is therefore defined as a microorganism capable of inducing the tissue destruction of apical periodontitis. This raises the question as to whether all microorganisms that inhabit the root canal space cause apical periodontitis, or whether specific organisms of the microbial flora are considered to cause the disease. Teeth with apical periodontitis almost always contain a polymicrobial infection and, whilst individual species may play different roles or dominate various stages of the infection, there is no evidence to suggest that particular species established therein are not involved in the pathogenesis of apical periodontitis.
Limited, but valuable information is available from a classic study (11) with an ‘eight-strain collection’ of species derived from one infected root canal that was reinoculated in equal proportions into other monkey teeth, either in toto or in various combinations of several or separate species. At the end of the experimental period, one species, Bacteroides oralis (now Prevotella oralis), dominated in mixed infections and demonstrated a more potent capacity for tissue destruction. In pure culture, however, B. oralis could not be re-isolated from inoculated root canals (11). Such findings illustrate the complexity of microbial interaction and that synergy between species may be essential for apical tissue destruction.
It is clear that much more work is needed to clarify the dynamics, functions and significance, individually and collectively, of the species constituting the root canal flora. The habitat-selected microorganisms in the infected root canal are there because of what they share in biochemical and physiological characteristics and/or what they contribute to others by mutual synergy. As far as we know today, all bacteria that establish in a root canal can be considered endodontic pathogens.
The number of microorganisms in a root canal infection is of relevance to the outcome of the disease process. Detection of just 10–50 cells of a species in a sample with 106 total cells implies a limited number of that species in the root canal and the contribution, therefore, of that number of cells to the development of disease is likely to be low. This may be a reason why studies using molecular approaches have generally failed to disclose an association between the presence of symptoms and particular species (50, 51). Molecular techniques such as polymerase-chain-reaction-based methods (PCR) do not give information on the abundance of microorganisms, whereas culturing and serial dilution provides information about the number present in a sample, and therefore what contribution they can really make to apical periodontitis. Thus, isolation and cultivation of microorganisms remain essential tools for the analysis of pathogenicity of the root canal flora.
Challenges faced by microbes establishing in the root canal
For bacteria to establish successfully as endodontic pathogens, they must overcome a series of consecutive barriers on and within the host tooth. It is likely that bacteria act in concert with others to penetrate hardand soft-tissue barriers, establish and then survive in the root canal.
The progressive challenges that microbes face in establishing in the root canal are shown in Fig. 1. In the untreated canal, microbes must breach tooth enamel by entering via microleakage, cracks or caries then reach the pulp by invading dentine. The pulp has its own host defense mechanism, which must be overwhelmed by the collective microbial consortium. Once the pulp has been invaded, the microbes must acquire suitable nutrient sources and possibly compete with others for available nutrition. The nutrition available at the tooth surface may be very different from that in pulp or in necrotic material. Finally, microbes face potential killing by the host defense. Ultimately, if successful, microbes will induce an inflammatory response at the apex resulting in apical periodontitis (Fig. 1).
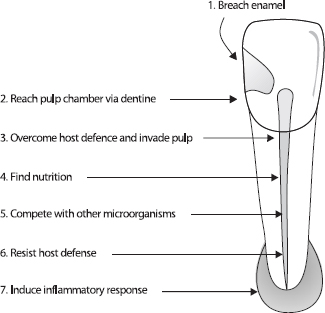
Fig. 1. Challenges for microbes to establish in the untreated root canal.
Comparison of culturing with molecular methods
Almost 40 years ago, Möller’s milestone publication (46) described how to sample the root canal in an accurate way, without contamination. The simple aim was to isolate and cultivate authentic microbial inhabitants of the root canal. With few exceptions, the papers published prior to that time had not applied the careful approach necessary for accurate isolation and cultivation. By documenting reproducible steps, recovery of microorganisms became possible without contamination from adjacent sites. The exacting principles applied to culture methods 40 years ago are no less important for recovery of microorganisms using the molecular technologies of today.
Acquiring accurate samples
Isolation of the tooth with rubber dam and decontamination of the rubber dam and tooth surface makes it possible to sample the root canal without contamination from the oral cavity (46). To check that the decontamination steps have been effective, control samples are required to ensure reliable sterility of the operative field prior to entry into the root canal (46). If the result of the control sample is positive, or in the absence of appropriate sterility checks, the veracity of the data from sampling may be in question because it is unknown whether the isolated bacteria are from within or outside the root canal.
Teeth should first be mechanically cleaned by polishing with pumice, then chemically cleaned and disinfected; before the sample is taken, the disinfectant should be inactivated so that it will not interfere with bacteria recovered from the root canal. During the access preparation, it is important to include a second decontamination stage and take a control sample before the root canal is reached, because removal of temporary fillings, carious dentin and bacteria under fillings (52) may contaminate the field (43, 46, 47). For sampling by culture, chemical cleansing with hydrogen peroxide is followed by disinfection with iodine (46).
Avoiding and managing contamination
Sampling and processing should provide a true representation of the root canal contents. When appropriate precautions are not applied, there is a risk that the results reported might not be authentic. The high sensitivity of molecular techniques, especially PCR, means that studies using these methods must use specific precautions to eliminate microbial DNA from the operation field during access preparation. When root canals are sampled by PCR, it has recently been shown that the method for decontamination requires modification (47). Cleansing with sodium hypochlorite is more efficient than hydrogen peroxide and iodine at removing detectable DNA from the tooth surface –13% of tooth surfaces were positive for detectable DNA, compared with 45%, respectively (47).
Contamination may occur not only at the site of sample recovery, but potentially during handling and laboratory procedures. The particular sensitivity of advanced culturing and molecular techniques means that many appropriate laboratory controls are necessary to avoid contamination and manage the risk. Culturing has the advantage that contamination during laboratory manipulation may be more readily recognized than with molecular procedures. PCR techniques have an inherently high risk that minute amounts of contaminating DNA are amplified and reported. There are many potential pitfalls of PCR-based analysis (53), so it is essential that the working environment is set up to reduce to a minimum the risk of contamination and that it is monitored with blank tubes, multiple positive and negative controls.
There are many procedural factors that should ensure the veracity of the results. Some examples are listed here to illustrate the kind of detailed attention necessary to deliver valid data:
- Primer design. Primers should be specific for the species and the amplified product sufficiently large. The specificity of the primers and their sensitivity should be tested and described. DNAases, released by some microorganisms at cell death, can degrade DNA in the material. The smaller fragments of DNA may persist for a longer time than larger sequences. Thus, designing the primers to target large, rather than small PCR products should yield fewer positive signals from dead cells.
- Cross-hybridization controls. There is a risk that the PCR primers designed from known 16S rRNA data may cross-hybridize with unrelated species. Therefore, controls should include checking by computer-based BLAST searches and cross-hybridization controls with live bacteria of related and unrelated species.
- Inhibition reactions. There is the possibility of inhibition reactions from constituents in the sample, which therefore require careful screening in clinical material. Collagen is one of many molecules known to be involved in inhibition reactions in clinical material.
- Optimizing PCR procedures. The optimal thermocycle parameters for PCR may differ between species. PCR conditions that are too, or insufficiently, stringent may result in poor PCR reactions with no, or wrong sequences amplified, respectively. The quality and quantity of extracted DNA depends on cell lysis conditions, which vary according to individual bacterial cell-wall structures. A low DNA recovery efficiency for some samples or some microorganisms may result in variable PCR results. For the method to be scientifically valid, it is important to verify the sensitivity and specificity of the primers.
- Sequencing the amplicon. After confirming that the amplicon is the predicted size, sequencing of the PCR product should substantiate that the amplified sequence matches that predicted from the expected species. The sequence match should be given along with the threshold for accepting identity match at species or genus levels.
Detection by PCR and culture
An important distinction needs to be made between what is evaluated by culture and PCR assays (38). Culturing measures viable bacterial cells as colony-forming units. Molecular methods measure nucleotide sequences and the PCR method allows amplification of very minute quantities of DNA to detectable levels.
Whether a root canal sample is evaluated by culturing or molecular techniques, microorganisms are acquired by the same method, usually by soaking up the fluid root canal contents with paper points. With this method, the results of laboratory processing depend on what can be recovered from the paper point. An inherent assumption in root canal sampling is that microorganisms acquired by paper point sampling reflect the type, number and diversity of the flora inhabiting the root canal. However, if the paper point does not reach all microbes, there remains a possibility that the sample may not accurately portray the root canal flora. The value and accuracy of the sample is critically dependent on how carefully the tooth is prepared, how scrupulously the sample is taken and the steps to exclude contamination. A contaminated sample is of little value regardless of whether it is later processed by molecular methods or culturing.
Culturing has the advantage that it allows all cells in a sample to grow, be subcultured and thus be identified. It has the limitation that some species have very stringent environmental and nutritional requirements that preclude culture on solid media; further, it is slow and time consuming. Culturing (and molecular methods) are highly technique sensitive and without appropriate checks may be prone to contamination and false results. Theoretically, a single bacterial cell can be detected by inoculating an undiluted root canal sample directly onto the sampling medium, but it is realistic to calculate that the limit of detection is 10–100 cells. There are two main reasons for this level of detection: serial dilution and cell survival.
Serial dilution of a sample is the primary reason for the detection limit of 10–100 cells. Serial dilution is required to:
- separate the bacteria into recognizable colonyforming units;
- reduce the numbers to 50–300 per plate for counting;
- distinguish different colony types for characterizing individual species.
During culturing some cells may die. This depends on the nutritional demands of the bacteria, how the sample is protected from oxygen exposure and on the inoculation time (54). The sensitivity of culturing can be improved with the use of selective media during subculture (38). Selective media can lower the detection limit not just because a particular species is promoted, but because other species are repressed, which then involves fewer dilutions to distinguish species.
PCR methods are well suited to target known or previously cultured and sequenced microorganisms. It can be done quickly with high specificity and where the target material is present in low amounts. PCR analysis has the advantage that it can amplify, without dilution, a particular sequence from the ambient background material. It is estimated that the DNA from 10 cells can be detected by PCR (38). Although PCR-based methods are highly specific, a disadvantage is that unidentified or non-targeted species cannot be detected, and cell numbers cannot be measured with conventional PCR. In 1996, a method that allows estimation of the number of bacteria, real-time PCR (RT-PCR), was described (55), but to our knowledge this method has not yet been applied in the analysis of endodontic infections.
Bacteria may be identified by PCR but not by culture (56, 57), although in some cases the difference is slight (58). Excluding the possibility of contamination, the likely reasons are the higher sensitivity of PCR and that PCR identifies DNA sequences, whether living or dead, compared with culturing that detects only viable cells. Whilst every living cell should be cultivable, bacteria may be undetectable by culture if (i) the number of cells is extremely low or (ii) bacteria are injured but not dead (59), (iii) the species is culture difficult (but possible) or (iv) the species is impossible to culture in vitro.
Limitations of PCR
Application of molecular methods in microbiology has revolutionized the taxonomic grouping of genera and allowed species to be systematically classified according to genetic structure rather than phenotypic behavior. The PCR technique in particular has allowed rapid identification of microorganisms that are difficult to culture. It is important, however, to recognize the limitations of this method so that interpretation of the data is reliable and valid.
The PCR method is a highly sensitive assay for DNA detection, but viable or dead cells are indistinguishable by their nucleotide sequence. Thus, microbial detection by PCR reveals current inhabitants of the root canal, but it may also represent a historical record of those microorganisms that have entered, but have not had the capacity to establish and survive. How long DNA from dead microorganisms may persist in the root canal is unknown. PCR methods have been successfully used to amplify Mycobacterium tuberculosis from several hundred to 1000-year-old animal and human remains (60–63).
The special binding affinity of hydroxyapatite for DNA has long been known (64) and this complicates recovery and analysis of the authentic endodontic microbial flora. Once DNA is adsorbed by dentine, it may prove difficult to extract (47, 65), and this problem raises several questions. For example, DNA from dead bacterial cells could be bound to coronal dentine, which could potentially contaminate the root canal sample. What is the fate of DNA from bacteria that have entered and not survived in the root canal? To date, these issues have not been adequately addressed in studies using molecular techniques, which leave open the question as to whether the reported species truly represent the living microbial flora of the root canal at the time of sampling.
There are few reports in the endodontic literature comparing results when alternative molecular methods have been applied to the same clinical material. In one study (66), checkerboard DNA–DNA hybridization was compared with 16S rDNA-based PCR. The two methods produced disparate results for the number of teeth positive for individual species and dissimilar matching between positive results. For example, Treponema denticola was detected by PCR in 23 of 50 samples and by checkerboard in five of the same 50 samples, with four samples positive by both methods. Tanerella forsythensis was positive in 11 of the 39 samples by PCR and in 15 samples by checkerboard, and only eight samples had matching positive results (66). Several factors, such as oligonucleotide design and preparation, and time/temperature protocols for hybridization and amplification could account for the disparate results.
The lack of studies with calibration of molecular methods suggests uncertainty about the authenticity and validity of reported results. Molecular methods offer sensitivity and precision, but a rigorous scientific approach with appropriate controls is essential for there to be confidence in the validity of the data.
Flora in untreated root canals
The root canal infection is a dynamic process and various bacterial species dominate at different stages of this process. In a long-standing infection, there is a shift towards dominance of the community by selected species. The most important factors driving this development are availability of nutrition, oxygen level (redox potential) and the local pH within the root canal.
Exogenous nutrients, such as fermentable carbohydrates, can affect the microbial ecology of the coronal parts of an exposed root canal, but endogenous proteins and glycoproteins are the principal nutrients in the main body of the root canal system. It might appear that the source of proteins in the root canal is restricted because of the progressive degradation of the small volume of pulpal tissue, but the bacteria induce a periapical inflammation that leads to influx of a serumlike exudate into the canal. This fluid is a sustainable nutrient source containing proteins and glycoproteins for those bacteria that have a proteolytic capacity. The bacteria that dominate this stage of the infection are likely to be those that either have a proteolytic capacity, or maintain a cooperative synergy with those that can utilize this substrate for bacterial metabolism. Bacterial metabolism of the serum-like fluid also causes reduction of the redox potential and a concomitant rise in the pH (8).
The species commonly recovered by culture from root canals of teeth with apical periodontitis have been previously reviewed (5). Application of molecular methods for microbial detection has meant that several additional species can be included as typical of the microbial flora of the infected root canal, which are described below.
Developments with molecular techniques
During the last decade, molecular techniques have increased our ability to differentiate bacteria and led to the establishment of new genera and species. To a great extent these are split off from previously established genera and species. An example of cultured bacteria that are subdivided and re-classified, are the fastidious, asaccharolytic, strict anaerobic, slow-growing, small, Gram-positive rods belonging to the genus Eubacterium. When they were first reported from root canal samples, the strains hydrolyzing arginine were characterized as Eubacterium lentum, and strains negative in this aspect were designated as ‘Eubacterium group 4’ (4). Later, strains belonging to the latter group were classified as a new species Eubacterium timidum (67). On the basis of phenotypic characteristics, DNA–DNA hybridization data and phylogenetic analysis with 16S rRNA gene sequence data, new species have been established from these two eubacteria. Oral strains of E. lentum have been re-classified as Eubacterium exiguum, later Slackia exigua (68, 69). The new genera and species Mogibacterium timidum, Mogibacterium pumilum, Mogibacterium vescum (70), Mogibacterium neglectum (71) and Cryptobacterium curtum (72) have been established from root canal and periodontal isolates of E. timidum (68).
Whilst molecular methods have helped to differentiate these species, the isolates were originally obtained by culturing from infected canals. A comparison between advanced culturing and molecular methods is possible for the species E. lentum and S. exigua, since S. exigua is the only species derived from E. lentum (68). With a species-specific PCR primer, and a detection limit of 10 organisms, S. exigua was found in 41% of teeth undergoing root canal treatment (73). This corresponds well with culturing where E. lentum has been found in 31% of teeth with necrotic pulps and periapical lesions (28). The material may not be fully comparable, but the differences in detection limits (approximately 0.5% for culture) may indicate that when this bacterium occurs, it is in numbers above this level.
Recently, studies using PCR have reported the species Dialister pneumosintes (formerly Bacteroides pneumosintes) and Filifactor alocis (formerly Fusobacterium alocis) to occur in 66% and 46% of root canals of teeth with apical periodontitis, respectively (74, 75). Because these species are non-fermentative, strictly anaerobic, slow-growing, forming diminutive colonies even after long incubation and unreactive in most biochemical tests, there are few reports of identification in culture isolates. Although D. pneumosintes was only described as a species in 1994 (76), they have been isolated by culturing (28, 77), but in lower frequencies (30–35%). The sensitivity of PCR-based screening is one obvious reason for the higher frequency of detection, but the problems involved in growth, isolation and identification are further possible reasons for infrequent identification by culture methods.
Using the PCR method, several bacterial species have been found to be more prevalent in root canals than previously reported by culture-based methods and a number of selected species are shown in Table 1.
Black-pigmented bacteria
Black-pigmented bacteria (BPB) have frequently been isolated from infected
root canals and, due to their proteolytic activity, are implicated in apical abscess formation (18, 20, 21, 78, 79). P. intermedia (formerly Bacteroides intermedius) has
been the most commonly isolated species. In 1992, isolates previously classified as P.
intermedia were re-classified as a new species, P. nigrescens (80). P. intermedia and P. nigrescens cannot be differentiated
using routine phenotypic methods. Using sodium dodecyl sulfate–polyacrylamide gel
electrophoresis and PCR, it has since been shown that P. nigrescens is more common in
endodontic infections than P. intermedia (81). P. intermedia and
P. nigrescens have been cultured from 26–40% of root canals of teeth with apical
periodontitis (28, 32, 82), although in
one PCR-based study, P. intermedia/nigrescens was detected in only 13% of infected
root canals (51).
Table 1. Comparison of culture (28) and molecular methods – selected species from untreated infected root canals
Prevalence (%)
| Study | F. nucleatum | Streptococcus sp.* | P. micros | P. propionicum | A. israelii | P. alactolyticus | P. intermedia/ P. nigrescens |
P. gingivalis | P. endodontalis | C. rectus | F. alocis | Enterococcus sp. |
| Fouad et al. (50) | 82 | 41 | 50 | 36 | 9 | 18 | 14 | |||||
| Siqueira et al. (102) | 10 | 5 | ||||||||||
| Siqueira et al. (51) | 13 | 28 | 43 | |||||||||
| Siqueira et al. (120) | 23 | 7 | 7 | |||||||||
| Siqueira & Rôças (209) | 23 | |||||||||||
| Siqueira & Rôças (210) | 56 | |||||||||||
| Siqueira & Rôças (198) | 36 | |||||||||||
| Siqueira & Rôças (74) | 46 | |||||||||||
| Sundqvist (28) | 48 | 35 | 34 | 8 | 11 | 34 | 34 | 5 | 9 | 25 | 29 | 2 |
| *Includes facultative and stricdly anaerobic species. |
The species Prevotella tannerae has also been included in the BPB-group since strains of this species may produce tan to black pigment when grown on rabbit blood agar (83). Root canal isolates initially characterized as P. intermedia/nigrescens (84) have then been shown to be P. tannerae by 16S rRNA gene sequencing (85). Interestingly, PCR using primers specific for P. tannerae revealed that 60% of clinical isolates from root canals and abscesses/cellulitis of endodontic origin were positive for P. tannerae. Thus, P. tannerae is an example of a species that was detected in earlier culturing studies, but under a former name. The higher frequency reported in molecular compared with culturing studies is partly explained by the inconsistency of this species to form pigmented colonies (83), so that isolates may have been classified as non-pigmenting saccharolytic Prevotella species.
Of other BPB, P. endodontalis and P. gingivalis have been reported, in culture studies, to occur in frequencies lower than 10% (28, 82). In contrast, PCR assays have detected P. endodontalis and P. gingivalis in 43% and 28%, respectively, of samples from necrotic pulps (51). The sensitivity of the PCR method probably accounts for the higher reported prevalence of Porphyromonas species.
Identification of culture-difficult species with molecular techniques
Spirochetes are the group of organisms for which PCR-based identification has brought about the greatest revision of reported prevalence in endodontic infections. More than 100 years ago, some of Miller’s drawings (9) clearly indicated the presence of spirochetes. Spirochetes have then been found in necrotic root canals using microbiological methods (7, 86, 87), dark-field microscopy (88–90) and transmission electron microscopy (91). Together, these publications suggested that spirochetes were only occasionally found and when they occurred, made up a small proportion of the flora.
Recently, evidence has emerged from PCR-based analysis that infected root canals contain a range of spirochetes in much higher prevalence than was previously thought (Table 2). The most predominant spirochetes in infected root canals are T. denticola (92–94) and Treponema socranskii (92, 93). The species Treponema lecithinolyticum (95) and Treponema maltophilum (92, 95, 96) are moderately prevalent, and Treponema amylovorum (92, 95), Treponema medium (95), Treponema pectinovorum (92, 93) and Treponema vincentii (92, 93) are infrequent inhabitants of the infected root canal (Table 2).
These studies (Table 2) show considerable diversity of values between research groups, for example prevalence of T. denticola has been reported at 13% and 78% (50, 93). Even the results derived from the same research group and clinical material reveal dissimilar prevalence values, for example T. denticola is reported at 43% and 78% (93, 94). An issue yet to be resolved is whether or not a relationship exists between the presence of spirochetes and symptoms. Some findings support such a relationship (92), whereas other data suggest no association (93). Despite some progress in understanding spirochete ecology and pathogenicity (97), more information is needed to explain their role in endodontic infections.
Tanerella forsythensis (formerly Bacteroides forsythus) is an example of a bacterium that is extremely difficult to culture. This Gram-negative anaerobe is dependent on other bacteria for growth and will not grow independently in vitro, if the medium is not supplemented with N-acetyl muramic acid (98). T. forsythensis is implicated in marginal periodontitis, but to our knowledge has not been cultured from root canals. This organism was first described in infected root canals when PCR-based tests revealed evidence of it in 18% of sampled root canals (99). Using similar methods, other groups have found evidence that T. forsythensis is a relatively frequent inhabitant (Table 3), with reported prevalence values of 16–55% (50, 66, 96, 100–103).
Molecular techniques – differentiation or new species?
Molecular methods have greatly facilitated identification of
culture-difficult species and expedited taxonomic grouping with ease and enhanced precision.
However, the impact of molecular methods on understanding the diversity of the root canal microbiota
has not been as dramatic as it might seem. With the exception of spirochetes and the species T.
forsythensis, which are prevalent in infected root canals yet difficult to cultivate,
molecular methods have identified species that have been previously detected by conventional
culture.
Table 2. Spirochetes in infected root canals
| Prevalence (%) | ||||||||||
| Study | No. of cases (n) |
T. amylovorum | T. denticota | T. lecithinolyticum | T. malthophilum | T. medium | T. pectinovorum | T. socranskii | T. vincentii | Other Treponemes |
| Baumgartner et al. (92) | 138 | 29 | 30 | 14 | 45 | 5 | 2 | |||
| Fouad et al. (50) | 24 | 13 | ||||||||
| Jung et al. (96) | 0 | 0 | 26 | 0 | 3 | 0 | ||||
| Rôças et al. (93) | 32 | 78 | 9 | 41 | 16 | |||||
| Siqueira & Rôças (95) | 31 | 7 | 26 | 39 | 13 | |||||
| Siqueira et al. (94) | 54 | 43 | ||||||||
Table 3. Tanerella forsythensis (formerly Bacteroides forsythus) in infected root canals
| Study | Number of cases (n) | Prevalence (%) |
| Fouad et al. (50) | 24 | 17 |
| Gonçalves & Mouton (100) | 11 | 55 |
| Jung et al. (96) | 73 | 16 |
| Rôças et al. (101) | 50 | 26 |
| Siqueira et al. (102) | 80 | 20 |
| Siqueira et al. (66) | 39 | 28 |
| Siqueira & Rôças (103) | 50 | 52 |
Flora in root-filled canals
It is generally acknowledged that persistence of disease is most commonly due to difficulties that occur during initial endodontic treatment. Inadequate aseptic control, poor access cavity design, missed canals, inadequate instrumentation, and leaking temporary or permanent restorations are examples of procedural pitfalls that may result in endodontic post-treatment disease (104).
The reasons for disease persistence in well-treated root-filled teeth have been poorly characterized until a series of studies published during the 1990’s. Using block biopsy material from non-healed periapical tissues including apices of the root-filled teeth, analysis by correlative light and electron microscopy has shown that there are five factors that may contribute to persistence of a periapical radiolucency after treatment. The factors are: (i) intraradicular infection (105); (ii) extraradicular infection by bacteria of the species Actinomyces israelii and Propionibacterium propionicum (106–108); (iii) foreign body reaction (109, 110); (iv) cysts, especially those containing cholesterol crystals (111); and (v) fibrous scar tissue healing (112). Of all these factors, it is generally believed that the major cause of persistent disease after root canal treatment is the persistence of microorganisms in the apical part of root-filled teeth.
Endodontic post-treatment disease, or apical periodontitis associated with a root-filled tooth, can continue for many years and may become apparent only when a tooth requires a new restoration. The fact that some microorganisms are capable of survival under harsh, nutrient-limited conditions of the root-filled canal for so long is remarkable. Yet, little information was known about the microorganisms involved in persistent intracanal infection after root filling until 1998, when two studies revealed that the microbial flora associated with endodontic post-treatment disease is quite unlike that found in other oral infections, or that of the untreated root canal (44, 45).
Microbiology of canals with persistent infection
Usually one or just a few species are recovered from canals of teeth with post-treatment disease. These are predominantly Gram-positive microorganisms and there is an equal distribution of facultative and obligate anaerobes (44, 45). This microbial flora is distinctly different from infections in untreated root canals, where the latter typically consists of a polymicrobial mix with approximately equal proportions of Grampositive and Gram-negative species, dominated by obligate anaerobes.
There is some diversity of species isolated from rootfilled teeth with persistent periapical disease, but there is a consensus amongst most studies that there is a high prevalence of enterococci and streptococci (44–46, 113–117). Other species found in higher proportions in individual studies are lactobacilli (44), Actinomyces species and peptostreptococci (116) and P. alactolyticus, P. propionicum, D. pneumosintes, and F. alocis (113). Some bacteriological findings from studies of root-filled teeth with persistent disease are shown in Table 4.
There is a difference in the microbial flora between poorly treated and
well-treated teeth when the canals are sampled at re-treatment. Although only one
poorly root-filled tooth was reported, the polymicrobial flora was found to be similar to
that seen in untreated root canals (45). This observation has recently been
confirmed in a study (117) where comparison of the isolates in 38 poorly filled
canals with 22 well-filled canals revealed a significant association of the former with
polymicrobial infections. When teeth are poorly treated, it is not surprising that the flora after
root canal filling should approximate that of the untreated canal, especially if it is also poorly
restored and there is microleakage from the oral cavity that allows an influx of carbohydrates and
possibly new bacteria.
Table 4. Bacteriological findings in root filled teeth with persistent periapical lesions
| Study | Species per root canal with bacteria | Enterococcus sp.* | Streptococcus sp.* | Candida sp.* | Actinomyces sp.* |
| Möller (46) | 1.6 | 29 | 16 | 3 | ND |
| Molander et al. (44) | 1.7 | 47 | 20 | 4 | 3 |
| Sundqvist et al. (45) | 1.3 | 38 | 25 | 8 | 13 |
| Hancock et al. (116) | 1.7 | 32 | 21 | 3 | 27 |
| Peciuliene et al. (115) | 1.6 | 64 | - | 18 | - |
| Cheung & Ho (118) | 2.6(1.8)‡ | ND | 50 | 17 | ND |
| Pinheiro et al. (117) | 2.1(1.8)‡ | 55 | 33 | 4 | 20 |
| Siqueira & Rôças (113)† | 4.1 | 77 | 23 | 9 | 5 |
*Percent prevalence, in canals with microorganisms.
†Identification by PCR. All other studies by culture.
‡Excluding poorly filled root canals.
ND, not detected.
The prevalence of enterococci has been a conspicuous finding in all studies that have investigated the flora in root-filled teeth (44–46, 113–117), with one exception (118), and implicates Enterococcus faecalis as an opportunistic pathogen in persistent apical periodontitis. Streptococci are also commonly isolated from root-filled canals with persistent lesions (Table 4). Other microorganisms of interest because of their association with endodontic post-treatment disease are species of Actinomyces and Candida. Some properties of these species are described in more detail below.
Enterococci
Studies that have recovered microbes from filled root canals with persistent periapical disease have shown a high proportion of enterococci, ranging from 29% to 77% (44– 46, 113, 115–117). This contrasts with a rather low proportion of enterococci, around 5% or less, recovered from untreated infected root canals (5–7, 119, 120) and raises the question of how and when enterococci establish in the root canal. Although more research is needed to address this issue, there are several possible explanations.
One possibility is that E. faecalis could be present in untreated canals, but in such low numbers that it is not recovered, or is outcompeted by other microorganisms in the bacterial consortium. When environmental conditions improve, it may grow to higher and recoverable proportions. In animal experiments (11), after inoculation of an eight-strain collection in equal (12.5%) proportions, E. faecalis was re-isolated at about 1% of the total microbial flora, which was similar to its proportion when originally recovered from an infected tooth. Whilst this might account for some cases, it is unlikely to explain all cases since even with sensitive molecular methods, E. faecalis was only detected in 7.5% of infected root canal samples (120) compared with ten times that prevalence in canals associated with post-treatment disease (113).
There must be another explanation for the high prevalence of E. faecalis in root-filled canals associated with disease and the most likely one is that E. faecalis enters the canal in the process of treatment, during or between treatment sessions. E. faecalis has been found in a higher proportion of canals that were inadequately sealed for a period of time during the treatment, or were treated over 10 or more sessions (121). Although it is unlikely to occur when the tooth has been wellrestored, it is conceivable that E. faecalis could enter after root filling, as it has been shown that poorly restored teeth have a higher rate of endodontic posttreatment disease (122).
Enterococci are part of a stable host-adapted bacterial community inhabiting the large bowel of most adult humans in numbers as high as 108 cfu/g of feces (123). They have a commensal relationship with the host, but under favorable circumstances may take advantage of temporary weaknesses in the host defense to establish infection. The species E. faecalis has some intrinsic characteristics that allow it to survive in conditions that are commonly lethal for many other microorganisms. These properties include an ability to grow in high salt concentrations (6.5% NaCl), a wide temperature range (10–60°C), 40% bile, a broad pH range, as well as persist in the presence of detergents (124–129).
E. faecalis and Enterococcus faecium are significant human pathogens particularly in nosocomial and antibiotic-resistant infections, yet their virulence factors are just beginning to be understood (130–135). Some virulence factors identified to date (123) are: (i) secreted factors such as a cytolysin and gelatinase (136); (ii) adhesins such as aggregation substance, enterococcal surface protein (Esp), collagen adhesin (Ace) (137– 141); and (iii) surface structures such as capsular polysaccharide (142). A notable cause for concern has been the special capacity of E. faecalis for acquiring antibiotic resistance genes from other organisms or by spontaneous mutation, making it particularly difficult to control an established enterococcal infection (143).
The characteristics required for persistent infection in the root canal are unlikely to be the same as those seen in soft-tissue infection in other parts of the body. One pathogenic property is a special capacity for invasion of dentinal tubules (144–146), particularly in the presence of serum (15) and in the absence of immunoglobulin G (147). Adhesion of E. faecalis to dentine could be another factor of relevance for pathogenesis. A recent study has shown that the serine protease and a collagen-binding protein (Ace) are involved in binding E. faecalis to dentine (148).
The intrinsic capacity of E. faecalis to withstand a wide pH range represents a problem for clinical antibacterial control. Calcium hydroxide, which is generally a highly potent antimicrobial dressing (40, 149, 150), is ineffective because E. faecalis can endure a high alkalinity up to around pH 11.5 (40, 145, 146, 151–154). The natural buffering effect of dentine (155–158) affords further protection to alkaline-resistant organisms since levels in dentine do not reach higher than pH 10.8 in cervical and pH 9.7 in apical dentine (156). The mechanisms of alkaline tolerance in E. faecalis have been essentially unknown until recently when it was shown that a functioning cell-wall-associated proton pump, which drives protons into the cell to acidify the cytoplasm, is important for survival of E. faecalis in a highly alkaline environment (151). Whilst the ability of E. faecalis to resist the antimicrobial effect of calcium hydroxide remains a significant clinical challenge in endodontic re-treatment, it may not be a critical factor for its involvement in post-treatment disease. A recent study of re-treated teeth in a North American population, where calcium hydroxide is infrequently used as a root canal dressing, showed that E. faecalis was recovered in similarly high proportions (116), which suggests that resistance to calcium hydroxide may not be the explanation for selection of this microorganism.
Another inherent characteristic of enterococci is an ability to adapt to fluctuating levels of nutrient supply and limitation, and it is this trait that may facilitate the persistence of E. faecalis in the canal long after root filling. Recently, this property was explored in a series of long-term starvation assays (159). E. faecalis survived in water for more than 4 months, which demonstrated the capacity of E. faecalis to endure long-term starvation. At the onset of starvation there was a rapid fall in viable cell numbers, leaving a residual small population of starved cells (159). These starvationstate cells were shown to be in a minimal metabolic state, since addition of cell-wall and DNA synthesis inhibitors to E. faecalis starvation cultures resulted in limited change in the rate of loss of viable cell numbers.
Although there is little known about the source and type of nutrition available at the apex of a root-filled canal, the microbial flora may be sustained by a periapical tissue transudate. This is likely to be a serum-derived fluid from surrounding tissue (15, 160). Growth of E. faecalis in serum is possible (15, 161, 162). Long-term experiments with cultures of E. faecalis in human serum showed a high number of cells were still viable after 4 months (159). Cells already in a starvation state were shown to be capable of recovery upon addition of serum (159). It is likely that E. faecalis may encounter periods of starvation in the root-filled canal, broken by opportunities to access serum or serum-like fluid. Under such conditions, even a small number of cells can gain the nutritional support required for survival and would therefore have the potential to maintain a periapical disease process.
A more detailed review of enterococci and their role in post-treatment apical periodontitis appears elsewhere in this issue.
Streptococci
Streptococci comprise a relatively high proportion, approximately 20% (range 16–50%, Table 4), of the microorganisms recovered from the canals of teeth with post-treatment disease (44, 45, 115–118). However, the recovery of streptococci is less remarkable when it is taken in the context of its high prevalence in untreated infected canals (5, 32).
The genus Streptococcus contains a diverse range of species of which oral streptococci fall into four broad groups (163, 164). Analysis of the Streptococcus species isolated from teeth with endodontic post-treatment disease indicates that no particular species or group have a higher prevalence. What streptococci have in common is a preferential capacity for invasion of dentinal tubules (165–167), which should favor their ability to enter and establish in the root canal system. Streptococcal surface adhesins mediate binding to dentin as well as facilitating dentin invasion (166, 167) and streptococcal invasion of dentin may also facilitate co-invasion of other species (168).
The ability of streptococci to penetrate or hide in dentinal tubules may be attributable to their pattern of growth in chains, a phenotypic characteristic shared with enterococci. This ability may also account for the finding of streptococci in approximately the same prevalence in initial and post-treatment root canal infections.
There is some evidence suggesting that streptococci are difficult to eradicate during treatment of the root canal. In a study that evaluated bacteria before and after instrumentation of the root canals, Streptococcus species were repeatedly isolated at up to three sessions of treatment (32). Interestingly, in the same study, Candida species were also difficult to eradicate, which demonstrates the challenges faced in antimicrobial control.
Candida
Candida albicans has been periodically reported in teeth with persistent post-treatment apical periodontitis (44, 45, 113, 115–118) and yeasts have also been observed by electron microscopy in such teeth (105). Yeasts are seldom seen in untreated root canals, unless canals have been open to the oral cavity (169) or there has been a history of protracted treatment (170). In one study, the prevalence of C. albicans in infected root canals was reported to be higher, although the type of clinical material was not stated (171).
Yeasts have several properties in common with enterococci. Yeasts have the capacity to survive as a monoinfection (170, 172) and several studies have shown a capacity for growth and invasion of dentine (173–175), although in comparison with E. faecalis, this property is weak (175). Not surprisingly, sodium hypochlorite is a potent agent in killing Candida species (176–178) and EDTA is also reported to be effective (179). Several in vitro studies have reported that Candida species resist the antimicrobial action of calcium hydroxide (176, 180), which may be a factor for selection of Candida in persistent root canal infections.
These characteristics suggest that both Candida and enterococci share several properties necessary to establish and survive in the harsh environment of the rootfilled canal. The properties include resistance to antimicrobials used in endodontic treatment, an ability to grow in monoinfections, survival in conditions of nutrient limitation and an ability to evade the host response by sequestration within the root canal system.
Actinomyces
A. israelii is of interest because it is a known and repeated culprit in therapy-resistant cases (107, 181– 183) and is by far the most common species involved in actinomycosis (184). The likely site of A. israelii infection is the periapical tissues where it is known to be involved in periapical actinomycosis; however, it is interesting that it has been recovered from the root canals of re-treated teeth (45, 116, 117). The presence of A. israelii in the root canal suggests the possibility of a communication between the periapical tissues and the canal, where some protection may be afforded from the host defense.
How A. israelii establishes in the periapical tissues is unknown. It may grow out as a clump from the root canal into the periapical tissues, or it may be forced from the root canal during instrumentation, thus inoculating the periapical tissue. Studies of experimental infection with A. israelii in animals have shown characteristic lesions of a cohesive bacterial mass of branching filaments surrounded by host leukocytes (185–188).
Identification of Actinomyces species has been hampered by problems with traditional biochemical methods of characterization. Although some studies have applied DNA hybridization methods (120, 189–191), these are not readily applicable and reproducible from one lab to another. The partial characterization of the 16S rRNA gene (192) has facilitated the development of probes suited to widespread application (193–195).
A. israelii is the most prevalent Actinomyces species isolated from human abscesses; however, Actinomyces gerencseriae (formerly A. israelii serotype II) is also prevalent and they are found in 56% and 25% of human abscesses, respectively (184). Using checkerboard DNA–DNA hybridization analysis of root canal samples from teeth diagnosed with periapical abscesses, A. israelii and A. gerencseriae have been reported in 14.8% and 7.4% of samples, respectively (120); however, the role of A. gerencseriae in persistent infection after root filling is unknown.
Recently, a new Actinomyces species, Actinomyces radicidentis (196), was found to be involved in posttreatment disease (197). Using PCR-based detection, it has been shown to be present in untreated root canal infections and root-filled teeth with chronic apical periodontitis (198), although its prevalence in both types of infection was low.
Actinomyces species share some properties with enterococci, streptococci and Candida including a growth pattern of cohesive filaments or chains, resistance to antimicrobials used in endodontic treatment, an ability to grow in monoinfections and to evade the host response.
A more detailed review of Actinomyces species and their role in post-treatment apical periodontitis appears elsewhere in this issue.
Ecological differences between untreated and root-filled root canals
The untreated infected root canal is an environment that provides microorganisms with nutritional diversity in a shifting pattern over time. The species that establish have typically invaded by caries, cracks or microleakage around fillings and they seek shelter, nutrition and a favorable habitat. Initially, there may be an influx of carbohydrates facilitating growth of facultative anaerobes, but as the infection matures, the available nutrients are mainly peptides and amino acids, which favor anaerobic proteolytic species.
Whilst the microbial flora in an untreated infected root canal may experience feast, in the well-filled root canal there is predominantly famine. Most or all of the original necrotic pulp will have been eliminated leaving dry, barren conditions for surviving microbial cells. These microbes would experience a static environment and starvation, but with some luck may encounter a serum-like fluid transudate from the periapical tissue. The species that persist here are those that have either survived the antimicrobial treatment and are the last ones remaining, or have entered during treatment and found it possible to establish where others cannot do so.
Properties of species associated with endodontic post-treatment disease
With the exception of Actinomyces, which is primarily involved in extraradicular infection, other species associated with persistent intraradicular infection described here, i.e. Candida, streptococci and enterococci, can be viewed as opportunistic pathogens. A behavior in common is to leave their normal habitat of the oral cavity and establish elsewhere, in the root canal, where they take advantage of the local ecological change in the environment and where there has been elimination of microbial competitors.
For microbes to maintain apical periodontitis and cause post-treatment disease, they must do more than just survive in the root-filled canal; they must also possess the pathogenic properties necessary to perpetuate inflammation external to the root canal system. In general, microorganisms involved in persistent infections implement one of three strategies to evade the immune response – sequestration, cellular or humoral evasion (199). Sequestration involves a physical barrier between the microbe and the host. Cellular evasion means that microorganisms avoid leukocyte-dependent antibacterial mechanisms. Humoral evasion means that extracellular bacteria avoid the host’s antibodies and complement.
At least two of the three strategies are deployed by microorganisms involved in endodontic post-treatment disease (200). A. israelii is an example of an endodontic pathogen that displays cellular evasion by avoiding phagocytosis by PMN leukocytes in vivo (185, 187, 188) primarily through a mechanism of collective cohesion (188). E. faecalis and Candida species are representative of microbes that are able to remain sequestered within the root canal system.
The properties necessary for microorganisms to persist in the root-filled canal are outlined in Fig. 2. Some of the physiological traits required for entry and initial establishment may be similar to that of microbes inhabiting a necrotic pulp in an untreated canal, such as an ability to find nutrients, compete with other microorganisms and evade initial host defenses.
For species to survive endodontic treatment (Fig. 2, phase 2), there must be an ability to withstand biomechanical cleaning and antimicrobial dressing. There are numerous reports confirming the bactericidal efficacy of sodium hypochlorite against several species involved in persistent infection such as A. israelii (201), E. faecalis (151, 202, 203) and Candida (176–178). It therefore seems reasonable to assume that these species may have the capacity to shelter from the main root canal in web-like areas, or in dentinal tubules where some level of protection or buffering of the antimicrobial agent is possible (157, 204). Although most root canal bacteria are sensitive to the high pH of calcium hydroxide (40), several species involved in persistent infection are now known to have a capacity to resist the antimicrobial effect of this commonly used agent (40, 145, 146, 151, 152, 180, 201).
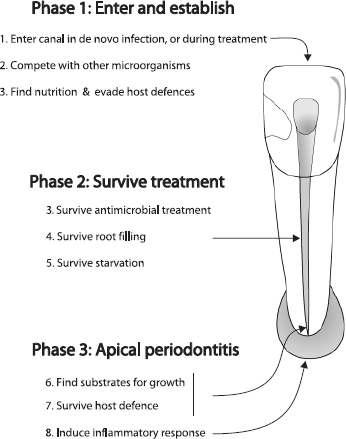
Fig. 2. Challenges for microbes involved in persistent infection.
How bacteria endure root filling is unknown, but studies that have sampled the root canal prior to root filling and then followed the treatment outcome of infected teeth have shown that some lesions heal (41, 45, 205, 206), implying that the bacteria did not survive or were not in a position to inflame the periapical tissue. Whether or not bacteria survive root canal filling may depend on whether they are entombed, or blocked from acquiring nutrition (104). It is possible, even likely, that bacteria may undergo a period of starvation. Here, the ability of E. faecalis to withstand periods of starvation (159, 207, 208), is a trait that may be crucial for survival.
Apical periodontitis is a dynamic process involving an interaction between host and living bacteria, and the microbes need to find substrates for growth (Fig. 2, phase 3). In a well-instrumented root canal where necrotic pulp tissue has been removed and there is no communication with exogenous nutrients from the oral cavity, nutrition is likely to come from a periapical fluid transudate, which is probably serum-like in nature (15). An ability to utilize collagen within dentine may also be useful and there are indications that E. faecalis may have this property (15, 148). The process of acquiring substrates for growth probably involves enzymatic breakdown of serum and tissue molecules, and this property in combination with an ability to avoid the host defense induce an inflammatory response in the periapical tissue.
Concurrent conditions for persistent infection
In a study that examined the influence of infection at the time of root filling on the outcome of treatment (41), 68% of teeth, which were infected at root filling, healed after the treatment. Similar results have also been reported in other studies (45, 205, 206). Whilst infection at the time of root canal filling will adversely affect the outcome of treatment, the presence of a persistent pathogen, alone, is not sufficient for persistence of disease. There must be a set of conditions that occur in combination to result in persistence of endodontic disease. These conditions are shown conceptually in Fig. 3. A set of microbial characteristics, coinciding with a set of location parameters permits an interaction with the host that will determine whether there will be persistent apical periodontitis.
The set of conditions for microorganisms include an ability to evade the antimicrobial stages of treatment, ‘persistence’ characteristics such as a starvation-survival potential, and a capacity to inflame host tissue (Fig. 3). Location parameters are also important. Provided that microbes can enter and reach the apical area, they must be situated near the apical (or accessory) foramen and have an open communication for the free exchange of fluid, molecules and organisms in order to inflame periapical tissue. The intersection of all these conditions with the host defense results in persistence of disease.
Fate of bacteria that have entered the root canal but do not survive
All bacteria have the theoretical capacity to enter and establish in the root canal, but few do so. Some may enter dentine, but do not reach the root canal. Others may reach the root canal, but do not survive. The fate of those bacteria that enter and reach the root canal but cannot establish or survive is unknown; however, their cell contents presumably disintegrate or are degraded by other microorganisms.
The fate of DNA from dead species is also uncertain. There remains a possibility that after lysis, the DNA fragments from these cells might linger in the canal or be bound to dentine and if so, such minute amounts would conceivably be detected and amplified by PCR. The presence of intrinsic or exogenous DNAases would also influence how long the DNA would persist.
In the only experimental study known to us that has examined the role and fate of a known microbial collection (11), various known combinations of an eight-strain collection of indigenous oral bacteria were inoculated into monkey teeth. At the end of the experimental period, Bacteroides oralis (now Prevotella oralis) dominated in mixed infections, yet could not be re-isolated when they had been inoculated in pure culture. The fate of the bacteria that were inoculated initially, but were not detected at the end of the experimental period is a matter of speculation. Whilst the species presumably died, it is also possible that some cells survived but in such low numbers that were not detectable by culture.
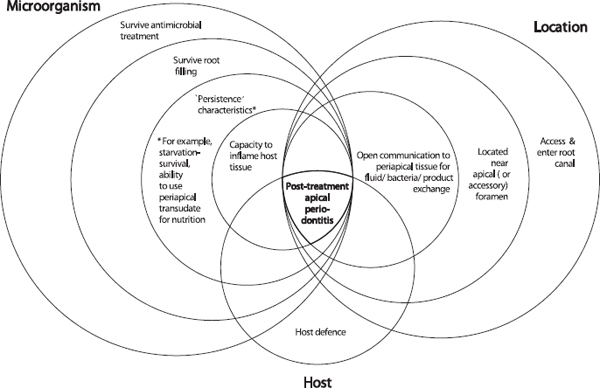
Fig. 3. Persistent infection requires not one, but a series of coupled characteristics. Bacteria must possess an ability to survive the stages of treatment, ‘persistence’ characteristics and an ultimate ability to inflame host tissue. The location of bacteria is critical for them to source nutrients and inflame tissue. The concomitant interaction of these characteristics with the host defense results in failure to heal.
In another study, subgingival plaque was grown in serum in a chemostat (16). One of the members of the microbial consortium, P. intermedia, was not detected initially, but after repeated serum enrichment it dominated the flora. This information shows that some bacteria can be present in low numbers, below the detection limit of the cultivation method.
Summary
Infection of the root canal is not a random event. The type and mix of the microbial flora develop in response to the surrounding environment. Factors that influence whether species die or survive are the particular ecological niche, nutrition, anaerobiosis, pH and competition or cooperation with other microorganisms. Species that establish a persistent root canal infection are selected by the phenotypic traits that they share in common and that are suited to the modified environment. Some of these shared characteristics include the capacity to penetrate and invade dentine, a growth pattern of chains or cohesive filaments, resistance to antimicrobials used in endodontic treatment, as well as an ability to grow in monoinfections, to survive periods of starvation and to evade the host response. Microorganisms that establish in the untreated root canal would experience an environment of nutritional diversity that changes with time. In contrast, the well-filled root canal offers the microbial flora little more than shelter from the host and microbial competitors, but in a small, dry, nutritionally limited space. In all cases, it is the environment that selects for microorganisms that possess traits suited to establishing and sustaining the disease process.
References
1. Mims C, Dimmock N, Nash A, Stephen J. Mims’ Pathogenesis of Infectious Disease. New York: Academic Press, 1995.
2. Moore WEC, Moore LVH. The bacteria of periodontal diseases. Periodontol 2000 1994: 5: 66–77.
3. Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol 2001: 183: 3770–3783.
4. Sundqvist G. Bacteriological studies of necrotic dental pulps. Odontological Dissertations No. 7. Department of Oral Microbiology, Umeå University, Sweden, 1976.
5. Sundqvist G. Taxonomy, ecology, and pathogenicity of the root canal flora. Oral Surg Oral Med Oral Pathol 1994: 78: 522–530.
6. Wittgow WC Jr, Sabiston CB Jr. Microorganisms from pulpal chambers of intact teeth with necrotic pulps. J Endod 1975: 1: 168–171.
7. Kantz WE, Henry CA. Isolation and classification of anaerobic bacteria from intact pulp chambers of nonvital teeth in man. Archs Oral Biol 1974: 19: 91–96.
8. Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology 2003: 149: 279–294.
9. Miller WD. An introduction to the study of the bacterio-pathology of the dental pulp. Dent Cosmos 1894: 36: 505–527.
10. Fabricius L, Dahlén G, Öhman AE, Möller ÅJR. Predominant indigenous oral bacteria isolated from infected root canals after varied times of closure. Scand J Dent Res 1982: 90: 134–144.
11. Fabricius L, Dahlén G, Holm SE, Möller ÅJR. Influence of combinations of oral bacteria on periapical tissues of monkeys. Scand J Dent Res 1982: 90: 200–206.
12. Loesche WJ, Gusberti F, Mettraux G, Higgins T, Syed S. Relationship between oxygen tension and subgingival bacterial flora in untreated human periodontal pockets. Infect Immun 1983: 42: 659–667.
13. Loesche WJ. Importance of nutrition in gingival crevice microbial ecology. Periodontics 1968: 6: 245–249.
14. Carlsson J, Frölander F, Sundqvist G. Oxygen tolerance of anaerobic bacteria isolated from necrotic dental pulps. Acta Odont Scand 1977: 35: 139–145.
15. Love RM. Enterococcus faecalis – a mechanism for its role in endodontic failure. Int Endod J 2001: 34: 399– 405.
16. ter Steeg PF, van der Hoeven JS. Development of periodontal microflora on human serum. Microb Ecol Health Dis 1989: 2: 1–10.
17. Persson S, Edlund MB, Claesson R, Carlsson J. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol 1990: 5: 195–201.
18. Sundqvist G, Carlsson J, Herrmann B, Tärnvik A. Degradation of human immunoglobulins G and M and complement factors C3 and C5 by black-pigmented Bacteroides. J Med Microbiol 1985: 19: 85–94.
19. Jansen H-J, van der Hoeven JS. Protein degradation by Prevotella intermedia and Actinomyces meyeri supports the growth of non-protein-cleaving oral bacteria in serum. J Clin Periodontol 1997: 24: 346–353.
20. Jansen H-J. The periodontal microflora as a proteindependent anaerobic degradation system. Department of Preventative Dentistry and Periodontology, University of Nijmegen, the Netherlands, 1996.
21. Sundqvist GK, Eckerbom MI, Larsson ÅP, Sjögren UT. Capacity of anaerobic bacteria from necrotic dental pulps to induce purulent infections. Infect Immun 1979: 25: 685–693.
22. Jansen H-J, van der Hoeven JS, Walji S, Goertz JH, Bakkeren JA. The importance of immunoglobulinbreakdown supporting the growth of bacteria in oral abscesses. J Clin Periodontol 1996: 23: 717–723.
23. Ohta H, Kato K, Fukui K, Gottschal JC. Microbial interactions and the development of periodontal disease. J Periodontal Res 1991: 26: 255–257.
24. Lev M, Keudell KC, Milford AF. Succinate as a growth factor for Bacteroides melaninogenicus. J Bacteriol 1971:108:175–178.
25. Marsh PD. Host defenses and microbial homeostasis: role of microbial interactions. J Dent Res 1989: 68: 1567–1575.
26. Grenier D, Mayrand D. Nutritional relationships between oral bacteria. Infect Immun 1986: 53: 616–620.
27. Gibbons RJ, Engle LP. Vitamin K compounds in bacteria that are obligate anaerobes. Science 1964: 146: 1307–1309.
28. Sundqvist G. Associations between microbial species in dental root canal infections. Oral Microbiol Immunol 1992: 7: 257–262.
29. Sundqvist G. Ecology of the root canal flora. J Endod 1992: 18: 427–430.
30. Carlsson J. Microbiology of plaque associated periodontal disease. In: Lindhe J, ed. Textbook of Clinical Periodontology. Copenhagen, Denmark: Munksgaard, 1990: 129–152.
31. Gomes BP, Drucker DB, Lilley JD. Positive and negative associations between bacterial species in dental root canals. Microbios 1994: 80: 231–243.
32. Lana MA, Ribeiro-Sobrinho AP, Stehling R, Garcia GD, Silva BK, Hamdan JS, Nicoli JR, Carvalho MA, de M. Farias L. Microorganisms isolated from root canals presenting necrotic pulp and their drug susceptibility in vitro. Oral Microbiol Immunol 2001: 16: 100–105.
33. Peters LB, Wesselink PR, van Winkelhoff AJ. Combinations of bacterial species in endodontic infections. Int Endod J 2002: 35: 698–702.
34. Kobayashi T, Hayashi A, Yoshikawa R, Okuda K, Hara K. The microbial flora from root canals and periodontal pockets of non-vital teeth associated with advanced periodontitis. Int Endod J 1990: 23: 100–106.
35. Moore WEC. Techniques for routine culture of fastidious anaerobes. Int J Syst Bacteriol 1966: 16: 173–190.
36. Hungate RE. The anaerobic mesophilic celluloytic bacteria. Bacteriol Rev 1950: 14: 1–49.
37. DiGuiseppi J, Fridovich I. The toxicology of molecular oxygen. Crit Rev Toxicol 1984: 12: 315–342.
38. Zambon JJ, Haraszthy VI. The laboratory diagnosis of periodontal infections. Periodontol 2000 1995: 7: 69–82.
39. Byström A, Sundqvist G. Bacteriologic evaluation of the efficacy of mechanical root canal instrumentation in endodontic therapy. Scand J Dent Res 1981: 89: 321– 328.
40. Byström A, Claesson R, Sundqvist G. The antibacterial effect of camphorated paramonochlorophenol, camphorated phenol and calcium hydroxide in the treatment of infected root canals. Endod Dent Traumatol 1985: 1: 170–175.
41. Sjögren U, Figdor D, Persson S, Sundqvist G. Influence of infection at the time of root filling on the outcome of endodontic treatment of teeth with apical periodontitis. (Published erratum appears in Int Endod J (1998) 31: 148). Int Endod J 1997: 30: 297–306.
42. Peters LB, van Winkelhoff AJ, Buijs JF, Wesselink PR. Effects of instrumentation, irrigation and dressing with calcium hydroxide on infection in pulpless teeth with periapical bone lesions. Int Endod J 2002: 35: 13–21.
43. Sundqvist G. Endodontic microbiology. In: Spångberg LSW, ed. Experimental Endodontics. Boca Raton, FL: CRC Press, 1990: 131–153.
44. Molander A, Reit C, Dahlén G, Kvist T. Microbiological status of root-filled teeth with apical periodontitis. Int Endod J 1998: 31: 1–7.
45. Sundqvist G, Figdor D, Persson S, Sjögren U. Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1998: 85: 86–93.
46. Möller ÅJR Microbiological examination of root canals and periapical tissues of human teeth. Methodological studies. Odontol Tidsk 1966: 74(Suppl): 1–380.
47. Ng YL, Spratt D, Sriskantharajah S, Gulabivala K. Evaluation of protocols for field decontamination before bacterial sampling of root canals for contemporary microbiology techniques. J Endod 2003: 29: 317–320.
48. Fors UG, Berg JO, Sandberg H. Microbiological investigation of saliva leakage between the rubber dam and tooth during endodontic treatment. J Endod 1986: 12: 396–399.
49. Casadevall A, Pirofski LA. Host–pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun 1999: 67: 3703–3713.
50. Fouad AF, Barry J, Caimano M, Clawson M, Zhu Q, Carver R, Hazlett K, Radolf JD. PCR-based identification of bacteria associated with endodontic infections. J Clin Microbiol 2002: 40: 3223–3231.
51. Siqueira JF Jr, Rôças IN, Oliveira JC, Santos KR. Molecular detection of black-pigmented bacteria in infections of endodontic origin. J Endod 2001: 27: 563–566.
52. Brännström M, Nyborg H. The presence of bacteria in cavities filled with silicate cement and composite resin materials. Swed Dent J 1971: 64: 149–155.
53. von Wintzingerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 1997: 21: 213–229.
54. Carlsson J, Sundqvist G. Evaluation of methods of transport and cultivation of bacterial specimens from infected dental root canals. Oral Surg Oral Med Oral Pathol 1980: 49: 451–454.
55. Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res 1996: 6: 986–994.
56. Rolph HJ, Lennon A, Riggio MP, Saunders WP, MacKenzie D, Coldero L, Bagg J. Molecular identification of microorganisms from endodontic infections. J Clin Microbiol 2001: 39: 3282–3289.
57. Munson MA, Pitt-Ford T, Chong B, Weightman A, Wade WG. Molecular and cultural analysis of the microflora associated with endodontic infections. J Dent Res 2002: 81: 761–766.
58. Martin FE, Nadkarni MA, Jacques NA, Hunter N. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J Clin Microbiol 2002: 40: 1698–1704.
59. Bogosian G, Bourneuf EV. A matter of bacterial life and death. EMBO Rep 2001: 2: 770–774.
60. Rothschild BM, Martin LD, Lev G, Bercovier H, Bar-Gal GK, Greenblatt C, Donoghue H, Spigelman M, Brittain D. Mycobacterium tuberculosis complex DNA from an extinct bison dated 17 000 years before the present. Clin Infect Dis 2001: 33: 305–311.
61. Salo WL, Aufderheide AC, Buikstra J, Holcomb TA. Identification of Mycobacterium tuberculosis DNA in a pre-Columbian Peruvian mummy. Proc Natl Acad Sci USA 1994: 91: 2091–2094.
62. Konomi N, Lebwohl E, Mowbray K, Tattersall I, Zhang D. Detection of mycobacterial DNA in Andean mummies. J Clin Microbiol 2002: 40: 4738–4740.
63. Fletcher HA, Donoghue HD, Holton J, Pap I, Spigelman M. Widespread occurrence of Mycobacterium tuberculosis DNA from 18th–19th century Hungarians. Am J Phys Anthropol 2003: 120: 144–152.
64. Bernardi G. Chromatography of nucleic acids on hydroxyapatite. Nature 1965: 206: 779–783.
65. Okazaki M, Yoshida Y, Yamaguchi S, Kaneno M, Elliott JC. Affinity binding phenomena of DNA onto apatite crystals. Biomaterials 2001: 22: 2459–2464.
66. Siqueira JF, Rôças IN, De Uzeda M, Colombo AP, Santos KR. Comparison of 16S rDNA-based PCR and checkerboard DNA–DNA hybridisation for detection of selected endodontic pathogens. J Med Microbiol 2002: 51: 1090–1096.
67. Holdeman LV, Cato EP, Burmeister EP, Moore WEC. Description of Eubacterium timidum sp. nov., Eubacterium brachy sp. nov. and Eubacterium nodatum sp. nov., isolated from human periodontitis. Int J Syst Bacteriol 1980: 30: 163–169.
68. Sato T, Sato M, Matsuyama J, Kalfas S, Sundqvist G, Hoshino E. Restriction fragment-length polymorphism analysis of 16S rDNA from oral asaccharolytic Eubacterium species amplified by polymerase chain reaction. Oral Microbiol Immunol 1998: 13: 23–29.
69. Poco SE Jr, Nakazawa F, Ikeda T, Sato M, Sato T, Hoshino E. Eubacterium exiguum sp. nov., isolated from human oral lesions. Int J Syst Bacteriol 1996: 46: 1120–1124.
70. Nakazawa F, Sato M, Poco SE, Hashimura T, Ikeda T, Kalfas S, Sundqvist G, Hoshino E. Description of Mogibacterium pumilum gen. nov., sp. nov. and Mogibacterium vescum gen. nov., sp. nov., and reclassification of Eubacterium timidum (Holdeman et al. 1980) as Mogibacterium timidum gen. nov., comb. nov. Int J Syst Evol Microbiol 2000: 50: 679–688.
71. Nakazawa F, Poco SE Jr, Sato M, Ikeda T, Kalfas S, Sundqvist G, Hoshino E. Taxonomic characterization of Mogibacterium diversum sp. nov. and Mogibacterium neglectum sp. nov., isolated from human oral cavities. Int J Syst Evol Microbiol 2002: 52: 115–122.
72. Nakazawa F, Poco SE, Ikeda T, Sato M, Kalfas S, Sundqvist G, Hoshino E. Cryptobacterium curtum gen. nov, sp. nov., a new genus of Gram-positive anaerobic rod isolated from human oral cavities. Int J Syst Bacteriol 1999: 49: 1193–1200.
73. Hashimura T, Sato M, Hoshino E. Detection of Slackia exigua, Mogibacterium timidum and Eubacterium saphenum from pulpal and periradicular samples using the Polymerase Chain Reaction (PCR) method. Int Endod J 2001: 34: 463–470.
74. Siqueira JF Jr, Rôças IN. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral Microbiol Immunol 2003: 18: 263–265.
75. Siqueira JF Jr, Rôças IN. Dialister pneumosintes can be a suspected endodontic pathogen. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2002: 94: 494–498.
76. Moore LV, Moore WE. Oribaculum catoniae gen. nov., sp. nov.; Catonella morbi gen. nov., sp. nov.; Hallella seregens gen. nov., sp. nov.; Johnsonella ignava gen. nov., sp. nov.; and Dialister pneumosintes gen. nov., comb. nov., nom. rev., Anaerobic gram-negative bacilli from the human gingival crevice. Int J Syst Bacteriol 1994: 44: 187–192.
77. Weiger R, Manncke B, Werner H, Löst C. Microbial flora of sinus tracts and root canals of non-vital teeth. Endod Dent Traumatol 1995: 11: 15–19.
78. van Winkelhoff AJ, Carlee AW, de Graaff J. Bacteroides endodontalis and other black-pigmented Bacteroides species in odontogenic abscesses. Infect Immun 1985: 49: 494–497.
79. Haapasalo M, Ranta H, Ranta K, Shah H. Blackpigmented Bacteroides spp. in human apical periodontitis. Infect Immun 1986: 53: 149–153.
80. Shah HN, Gharbia SE. Biochemical and chemical studies on strains designated Prevotella intermedia and proposal of a new pigmented species, Prevotella nigrescens sp. nov. Int J Syst Bacteriol 1992: 42: 542– 546.
81. Baumgartner JC, Bae KS, Xia T, Whitt J, David LL. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and polymerase chain reaction for differentiation of Prevotella intermedia and Prevotella nigrescens. J Endod 1999: 25: 324–328.
82. Wasfy MO, McMahon KT, Minah GE, Falkler WA Jr. Microbiological evaluation of periapical infections in Egypt. Oral Microbiol Immunol 1992: 7: 100– 105.
83. Moore LV, Johnson JL, Moore WE. Descriptions of Prevotella tannerae sp. nov. and Prevotella enoeca sp. nov. from the human gingival crevice and emendation of the description of Prevotella zoogleoformans. Int J Syst Bacteriol 1994: 44: 599–602.
84. Baumgartner JC, Watkins BJ, Bae KS, Xia T. Association of black-pigmented bacteria with endodontic infections. J Endod 1999: 25: 413–415.
85. Xia T, Baumgartner JC, David LL. Isolation and identification of Prevotella tannerae from endodontic infections. Oral Microbiol Immunol 2000: 15: 273– 275.
86. Hampp EG. Isolation and identification of spirochetes obtained from unexposed canals of pulp-involved teeth. Oral Surg 1957: 10: 1100–1104.
87. Dahle UR, Tronstad L, Olsen I. Characterization of new periodontal and endodontic isolates of spirochetes. Eur J Oral Sci 1996: 104: 41–47.
88. Brown LR, Rudolph CE. Isolation and identification of microorganisms from unexposed canals of pulp-involved teeth. Oral Surg 1957: 10: 1094–1099.
89. Thilo BE, Baehni P, Holz J. Dark-field observation of the bacterial distribution in root canals following pulp necrosis. J Endod 1986: 12: 202–205.
90. Dahle UR, Tronstad L, Olsen I. Observation of an unusually large spirochete in endodontic infection. Oral Microbiol Immunol 1993: 8: 251–253.
91. Nair PNR. Light and electron microscopic studies of root canal flora and periapical lesions. J Endod 1987: 13: 29–39.
92. Baumgartner JC, Khemaleelakul SU, Xia T. Identification of spirochetes (treponemes) in endodontic infections. J Endod 2003: 29: 794–797.
93. Rôças IN, Siqueira JF Jr, Andrade AF, Uzeda M. Oral treponemes in primary root canal infections as detected by nested PCR. Int Endod J 2003: 36: 20–26.
94. Siqueira JF Jr, Rôças IN, Favieri A, Oliveira JC, Santos KR. Polymerase chain reaction detection of Treponema denticola in endodontic infections within root canals. Int Endod J 2001: 34: 280–284.
95. Siqueira JF Jr, Rôças IN. PCR-based identification of Treponema maltophilum, T amylovorum, T medium, and T lecithinolyticum in primary root canal infections. Arch Oral Biol 2003: 48: 495–502.
96. Jung IY, Choi B, Kum KY, Yoo YJ, Yoon TC, Lee SJ, Lee CY. Identification of oral spirochetes at the species level and their association with other bacteria in endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001: 92: 329–334.
97. Chan EC, McLaughlin R. Taxonomy and virulence of oral spirochetes. Oral Microbiol Immunol 2000: 15: 1–9.
98. Wyss C. Dependence of proliferation of Bacteroides forsythus on exogenous N-acetylmuramic acid. Infect Immun 1989: 57: 1757–1759.
99. Conrads G, Gharbia SE, Gulabivala K, Lampert F, Shah HN. The use of a 16S rDNA directed PCR for the detection of endodontopathogenic bacteria. J Endod 1997: 23: 433–438.
100. Gonçalves RB, Mouton C. Molecular detection of Bacteroides forsythus in infected root canals. J Endod 1999: 25: 336–340.
101. Rôças IN, Siqueira JF Jr, Santos KR, Coelho AM. ‘Red complex’ (Bacteroides forsythus, Porphyromonas gingivalis, and Treponema denticola) in endodontic infections: a molecular approach. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001: 91: 468–471.
102. Siqueira JF Jr, Rôças IN, Moraes SR, Santos KR. Direct amplification of rRNA gene sequences for identification of selected oral pathogens in root canal infections. Int Endod J 2002: 35: 345–351.
103. Siqueira JF Jr, Rôças IN. Bacteroides forsythus in primary endodontic infections as detected by nested PCR. J Endod 2003: 29: 390–393.
104. Sundqvist G, Figdor D. Endodontic treatment of apical periodontitis;. In: Ørstavik D, Pitt Ford TR, eds. Essential Endodontology. Prevention and Treatment of Apical Periodontitis. Oxford, UK: Blackwell Science, 1998: 242–277.
105. Nair PNR, Sjögren U, Krey G, Kahnberg K-E, Sundqvist G. Intraradicular bacteria and fungi in rootfilled, asymptomatic human teeth with therapy-resistant periapical lesions: a long-term light and electron microscopic follow-up study. J Endod 1990: 16: 580–588.
106. Nair PNR, Schroeder HE. Periapical actinomycosis. J Endod 1984: 10: 567–570.
107. Sundqvist G, Reuterving C-O. Isolation of Actinomyces israelii from periapical lesion. J Endod 1980: 6: 602– 606.
108. Sjögren U, Happonen RP, Kahnberg KE, Sundqvist G. Survival of Arachnia propionica in periapical tissue. Int Endod J 1988: 21: 277–282.
109. Koppang HS, Koppang R, Solheim T, Aarnes H, Stolen SO. Cellulose fibers from endodontic paper points as an etiological factor in postendodontic periapical granulomas and cysts. J Endod 1989: 15: 369–372.
110. Nair PNR, Sjögren U, Krey G, Sundqvist G. Therapyresistant foreign body giant cell granuloma at the periapex of a root-filled human tooth. J Endod 1990: 16: 589–595.
111. Nair PNR, Sjögren U, Schumacher E, Sundqvist G. Radicular cyst affecting a root-filled human tooth: a long-term post-treatment follow-up. Int Endod J 1993: 26: 225–233.
112. Nair PNR, Sjögren U, Figdor D, Sundqvist G. Persistent periapical radiolucencies of root-filled human teeth, failed endodontic treatments, and periapical scars. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1999: 87: 617–627.
113. Siqueira JF Jr, Rôças IN. Polymerase chain reactionbased analysis of microorganisms associated with failed endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2004: 97: 85–94.
114. Peciuliene V, Balciuniene I, Eriksen HM, Haapasalo M. Isolation of Enterococcus faecalis in previously rootfilled canals in a Lithuanian population. J Endod 2000: 26: 593–595.
115. Peciuliene V, Reynaud AH, Balciuniene I, Haapasalo M. Isolation of yeasts and enteric bacteria in root-filled teeth with chronic apical periodontitis. Int Endod J 2001: 34: 429–434.
116. Hancock HH III, Sigurdsson A, Trope M, Moiseiwitsch J. Bacteria isolated after unsuccessful endodontic treatment in a North American population. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001: 91: 579–586.
117. Pinheiro ET, Gomes BP, Ferraz CC, Sousa EL, Teixeira FB, Souza-Filho FJ. Microorganisms from canals of root-filled teeth with periapical lesions. Int Endod J 2003: 36: 1–11.
118. Cheung GS, Ho MW. Microbial flora of root canaltreated teeth associated with asymptomatic periapical radiolucent lesions. Oral Microbiol Immunol 2001: 16: 332–337.
119. Byström A, Sundqvist G. The antibacterial action of sodium hypochlorite and EDTA in 60 cases of endodontic therapy. Int Endod J 1985: 18: 35–40.
120. Siqueira JF Jr, Rôças IN, Souto R, de Uzeda M, Colombo AP. Actinomyces species, streptococci, and Enterococcus faecalis in primary root canal infections. J Endod 2002: 28: 168–172.
121. Siren EK, Haapasalo MP, Ranta K, Salmi P, Kerosuo EN. Microbiological findings and clinical treatment procedures in endodontic cases selected for microbiological investigation. Int Endod J 1997: 30: 91–95.
122. Ray HA, Trope M. Periapical status of endodontically treated teeth in relation to the technical quality of the root filling and the coronal restoration. Int Endod J 1995: 28: 12–18.
123. Gilmore M, Coburn PS, Nallapareddy SR, Murray PR. Enterococcal virulence. In: Gilmore M, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB, eds. The Enterococci. Pathogenesis, Molecular Biology, and Antibiotic Resistance. Washington, DC, ASM Press, 2002: 301–354.
124. Hancock LE, Gilmore M. Pathogenicity of enterococci. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, eds. Gram-positive Pathogens. Washington, DC, ASM Press, 2000: 251–258.
125. Giard JC, Laplace JM, Rince A, Pichereau V, Benachour A, Leboeuf C, Flahaut S, Auffray Y, Hartke A. The stress proteome of Enterococcus faecalis. Electrophoresis 2001: 22: 2947–2954.
126. Boutibonnes P, Giard JC, Hartke A, Thammavongs B, Auffray Y. Characterization of the heat shock response in Enterococcus faecalis. Antonie Van Leeuwenhoek 1993: 64: 47–55.
127. Flahaut S, Hartke A, Giard JC, Benachour A, Boutibonnes P, Auffray Y. Relationship between stress response toward bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol Lett 1996: 138: 49–54.
128. Flahaut S, Hartke A, Giard JC, Auffray Y. Alkaline stress response in Enterococcus faecalis: adaptation, crossprotection, and changes in protein synthesis. Appl Environ Microbiol 1997: 63: 812–814.
129. Hartke A, Giard JC, Laplace JM, Auffray Y. Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl Environ Microbiol 1998: 64: 4238–4245.
130. Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev 1990: 3: 46–65.
131. Murray BE, Weinstock GM. Enterococci: new aspects of an old organism. Proc Assoc Am Physicians 1999: 111: 328–334.
132. Murray BE. Vancomycin-resistant enterococcal infections. N Engl J Med 2000: 342: 710–721.
133. Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clin Microbiol Rev 1994: 7: 462–478.
134. Huycke MM, Sahm DF, Gilmore MS. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis 1998: 4: 239– 249.
135. Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev 2000: 13: 686–707.
136. Mäkinen P-L, Clewell DB, An F, Mäkinen KK. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (‘gelatinase’) from Streptococcus faecalis (strain 0G1-10). J Biol Chem 1989: 264: 3325–3334.
137. Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun 2001: 69: 4366–4372.
138. Olmsted SB, Dunny GM, Erlandsen SL, Wells CL. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J Infect Dis 1994: 170: 1549–1556.
139. Nallapareddy SR, Singh KV, Duh RW, Weinstock GM, Murray BE. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of ace during human infections. Infect Immun 2000: 68: 5210–5217.
140. Schlievert PM, Gahr PJ, Assimacopoulos AP, Dinges MM, Stoehr JA, Harmala JW, Hirt H, Dunny GM. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect Immun 1998: 66: 218–223.
141. Rakita RM, Vanek NN, Jacques-Palaz K, Mee M, Mariscalco MM, Dunny GM, Snuggs M, Van Winkle WB, Simon SI. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun 1999: 67: 6067–6075.
142. Huebner J, Wang Y, Krueger WA, Madoff LC, Martirosian G, Boisot S, Goldmann DA, Kasper DL, Tzianabos AO, Pier GB. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect Immun 1999: 67: 1213–1219.
143. Mundy LM, Sahm DF, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev 2000: 13: 513–522.
144. Peters LB, Wesselink PR, Moorer WR. Penetration of bacteria in bovine root dentine in vitro. Int Endod J 2000: 33: 28–36.
145. Safavi KE, Spångberg LSW, Langeland K. Root canal dentinal tubule disinfection. J Endod 1990: 16: 207– 210.
146. Haapasalo M, Ørstavik D. In vitro infection and disinfection of dentinal tubules. J Dent Res 1987: 66: 1375–1379.
147. Love RM. The effect of tissue molecules on bacterial invasion of dentine. Oral Microbiol Immunol 2002: 17: 32–37.
148. Hubble TS, Hatton JF, Nallapareddy SR, Murray BE, Gillespie MJ. Influence of Enterococcus faecalis proteases and the collagen-binding protein, Ace, on adhesion to dentin. Oral Microbiol Immunol 2003: 18: 121–126.
149. Sjögren U, Figdor D, Spångberg L, Sundqvist G. The antimicrobial effect of calcium hydroxide as a shortterm intracanal dressing. Int Endod J 1991: 24: 119– 125.
150. Cvek M, Hollender L, Nord CE. Treatment of nonvital permanent incisors with calcium hydroxide. VI. A clinical, microbiological and radiological evaluation of treatment in one sitting of teeth with mature or immature root. Odontol Revy 1976: 27: 93–108.
151. Evans M, Davies JK, Sundqvist G, Figdor D. Mechanisms involved in the resistance of Enterococcus faecalis to calcium hydroxide. Int Endod J 2002: 35: 221–228.
152. Ørstavik D, Haapasalo M. Disinfection by endodontic irrigants and dressings of experimentally infected dentinal tubules. Endod Dent Traumatol 1990: 6: 142–149.
153. Tanriverdi F, Esener T, Erganiş O, Belli S. An in vitro test model for investigation of disinfection of dentinal tubules infected with Enterococcus faecalis. Braz Dent J 1997: 8: 67–72.
154. Siqueira JF Jr, de Uzeda M. Disinfection by calcium hydroxide pastes of dentinal tubules infected with two obligate and one facultative anaerobic bacteria. J Endod 1996: 22: 674–676.
155. Wang J-D, Hume WR. Diffusion of hydrogen ion and hydroxyl ion from various sources through dentine. Int Endod J 1988: 21: 17–26.
156. Nerwich A, Figdor D, Messer HH. pH changes in root dentin over a 4-week period following root canal dressing with calcium hydroxide. J Endod 1993: 19: 302–306.
157. Haapasalo HK, Sirén EK, Waltimo TM, Ørstavik D, Haapasalo MP. Inactivation of local root canal medicaments by dentine: an in vitro study. Int Endod J 2000: 33: 126–131.
158. Portenier I, Haapasalo H, Rye A, Waltimo T, Ørstavik D, Haapasalo M. Inactivation of root canal medicaments by dentine, hydroxylapatite and bovine serum albumin. Int Endod J 2001: 34: 184–188.
159. Figdor D, Davies JK, Sundqvist G. Starvation survival, growth and recovery of Enterococcus faecalis in human serum. Oral Microbiol Immunol 2003: 18: 234–239.
160. Morse DR, Patnik JW, Schacterle GR. Electrophoretic differentiation of radicular cysts and granulomas. Oral Surg 1973: 35: 249–264.
161. Guzmàn CA, Pruzzo C, Platé M, Guardati MC, Calegari L. Serum dependent expression of Enterococcus faecalis adhesins involved in the colonization of heart cells. Microb Pathog 1991: 11: 399–409.
162. Arduino RC, Murray BE, Rakita RM. Roles of antibodies and complement in phagocytic killing of enterococci. Infect Immun 1994: 62: 987–993.
163. Whiley RA, Beighton D. Current classification of the oral streptococci. Oral Microbiol Immunol 1998: 13: 195–216.
164. Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol 1995: 45: 406–408.
165. Perez F, Rochd T, Lodter JP, Calas P, Michel G. In vitro study of the penetration of three bacterial strains into root dentine. Oral Surg Oral Med Oral Pathol 1993: 76: 97–103.
166. Love RM, McMillan MD, Jenkinson HF. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect Immun 1997: 65: 5157– 5164.
167. Love RM, Jenkinson HF. Invasion of dentinal tubules by oral bacteria. Crit Rev Oral Biol Med 2002: 13: 171– 183.
168. Love RM, McMillan MD, Park Y, Jenkinson HF. Coinvasion of dentinal tubules by Porphyromonas gingivalis and Streptococcus gordonii depends upon binding specificity of streptococcal antigen I/II adhesin. Infect Immun 2000: 68: 1359–1365.
169. Sen BH, Piskin B, Demirci T. Observation of bacteria and fungi in infected root canals and dentinal tubules by SEM. Endod Dent Traumatol 1995: 11: 6–9.
170. Waltimo TM, Sirén EK, Torkko HL, Olsen I, Haapasalo MP. Fungi in therapy-resistant apical periodontitis. Int Endod J 1997:30: 96–101.
171. Baumgartner JC, Watts CM, Xia T. Occurrence of Candida albicans in infections of endodontic origin. J Endod 2000: 26: 695–698.
172. Waltimo TM, Sen BH, Meurman JH, Ørstavik D, Haapasalo MP. Yeasts in apical periodontitis. Crit Rev Oral Biol Med 2003: 14: 128–137.
173. Sen BH, Safavi KE, Spångberg LS. Growth patterns of Candida albicans in relation to radicular dentin. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1997: 84: 68–73.
174. Sen BH, Safavi KE, Spångberg LS. Colonization of Candida albicans on cleaned human dental hard tissues. Arch Oral Biol 1997: 42: 513–520.
175. Waltimo TM, Orstavik D, Siren EK, Haapasalo MP. In vitro yeast infection of human dentin. J Endod 2000: 26: 207–209.
176. Waltimo TM, Orstavik D, Siren EK, Haapasalo MP. In vitro susceptibility of Candida albicans to four disinfectants and their combinations. Int Endod J 1999: 32: 421–429.
177. D’Arcangelo C, Varvara G, De Fazio P. An evaluation of the action of different root canal irrigants on facultative aerobic-anaerobic, obligate anaerobic, and microaerophilic bacteria. J Endod 1999: 25: 351–353.
178. Sen BH, Safavi KE, Spångberg LS. Antifungal effects of sodium hypochlorite and chlorhexidine in root canals. J Endod 1999: 25: 235–238.
179. Sen BH, Akdeniz BG, Denizci AA. The effect of ethylenediamine-tetraacetic acid on Candida albicans. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2000: 90: 651–655.
180. Waltimo TM, Siren EK, Orstavik D, Haapasalo MP. Susceptibility of oral Candida species to calcium hydroxide in vitro. Int Endod J 1999: 32: 94–98.
181. Happonen R-P. Periapical actinomycosis: a follow-up study of 16 surgically treated cases. Endod Dent Traumatol 1986: 2: 205–209.
182. O’Grady JF, Reade PC. Periapical actinomycosis involving Actinomyces israelii. J Endod 1988: 14: 147–149.
183. Byström A, Happonen R-P, Sjögren U, Sundqvist G. Healing of periapical lesions of pulpless teeth after endodontic treatment with controlled asepsis. Endod Dent Traumatol 1987: 3: 58–63.
184. Schaal KP, Lee H-J. Actinomycete infections in humans–a review. Gene 1992: 115: 201–211.
185. Brown JR, von Lichtenberg F. Experimental actinomycosis in mice. Arch Path 1970: 90: 391–402.
186. Behbehani MJ, Jordan HV. Comparative pathogenicity of Actinomyces species in mice. J Med Microbiol 1982: 15: 465–473.
187. Sumita M, Hoshino E, Iwaku M. Experimental actinomycosis in mice induced by alginate gel particles containing Actinomyces israelii. Endod Dent Traumatol 1998: 14: 137–143.
188. Figdor D, Sjögren U, Sörlin S, Sundqvist G, Nair PNR. Pathogenicity of Actinomyces israelii and Arachnia propionica: experimental infection in guinea pigs and phagocytosis and intracellular killing by human polymorphonuclear leukocytes in vitro. Oral Microbiol Immunol 1992: 7: 129–136.
189. Ximénez-Fyvie LA, Haffajee AD, Martin L, Tanner A, Macuch P, Socransky SS. Identification of oral Actinomyces species using DNA probes. Oral Microbiol Immunol 1999: 14: 257–265.
190. Gatti JJ, Dobeck JM, Smith C, White RR, Socransky SS, Skobe Z. Bacteria of asymptomatic periradicular endodontic lesions identified by DNA–DNA hybridization. Endod Dent Traumatol 2000: 16: 197–204.
191. Sunde PT, Tronstad L, Eribe ER, Lind PO, Olsen I. Assessment of periradicular microbiota by DNA–DNA hybridization. Endod Dent Traumatol 2000: 16: 191–196.
192. Stackebrandt E, Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol 1990: 136: 37–43.
193. Tang G, Yip HK, Luo G, Cheung BP, Shen S, Samaranayake LP. Development of novel oligonucleotide probes for seven Actinomyces species and their utility in supragingival plaque analysis. Oral Dis 2003: 9: 203–209.
194. Jauh-Hsun C, Vinh T, Davies JK, Figdor D. Molecular approaches to the differentiation of Actinomyces species. Oral Microbiol Immunol 1999: 14: 250–256.
195. Sato T, Matsuyama J, Takahashi N, Sato M, Johnson J, Schachtele C, Hoshino E. Differentiation of oral Actinomyces species by 16S ribosomal DNA polymerase chain reaction-restriction fragment length polymorphism. Arch Oral Biol 1998: 43: 247–252.
196. Collins MD, Hoyles L, Kalfas S, Sundqvist G, Monsen T, Nikolaitchouk N, Falsen E. Characterization of Actinomyces isolates from infected root canals of teeth: description of Actinomyces radicidentis sp. nov. J Clin Microbiol 2000: 38: 3399–3403.
197. Kalfas S, Figdor D, Sundqvist G. A new bacterial species associated with failed endodontic treatment: identification and description of Actinomyces radicidentis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001: 92: 208–214.
198. Siqueira JF Jr, Rôças IN. Polymerase chain reaction detection of Propionibacterium propionicus and Actinomyces radicidentis in primary and persistent endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2003: 96: 215–222.
199. Nataro JP, Blaser MJ, Cunningham-Rundles S. Persistent bacterial infections: commensalism gone awry or adaptive niche? In: Nataro JP, Blaser MJ, Cunningham-Rundles S, eds. Persistent Bacterial Infections. Washington, DC, ASM Press, 2000: 3–10.
200. Figdor D. Microbial aetiology of endodontic treatment failure & pathogenic properties of selected species. Umeå University Odontological Dissertations No. 79. Department of Endodontics, Umeå University, Sweden, 2002.
201. Barnard D, Davies J, Figdor D. Susceptibility of Actinomyces israelii to antibiotics, sodium hypochlorite and calcium hydroxide. Int Endod J 1996: 29: 320– 326.
202. Siqueira JF Jr, Machado AG, Silveira RM, Lopes HP, De Uzeda M. Evaluation of the effectiveness of sodium hypochlorite used with three irrigation methods in the elimination of Enterococcus faecalis from the root canal, in vitro. Int Endod J 1997: 30: 279–282.
203. Spratt DA, Pratten J, Wilson M, Gulabivala K. An in vitro evaluation of the antimicrobial efficacy of irrigants on biofilms of root canal isolates. Int Endod J 2001: 34: 300–307.
204. Huque J, Kota K, Yamaga M, Iwaku M, Hoshino E. Bacterial eradication from root dentine by ultrasonic irrigation with sodium hypochlorite. Int Endod J 1998: 31: 242–250.
205. Trope M, Delano EO, Ørstavik D. Endodontic treatment of teeth with apical periodontitis: single vs. multivisit treatment. J Endod 1999: 25: 345–350.
206. Katebzadeh N, Sigurdsson A, Trope M. Radiographic evaluation of periapical healing after obturation of infected root canals: an in vivo study. Int Endod J 2000: 33: 60–66.
207. Giard JC, Hartke A, Flahaut S, Benachour A, Boutibonnes P, Auffray Y. Starvation-induced multiresistance in Enterococcus faecalis JH2-2. Curr Microbiol 1996: 32: 264–271.
208. Wendt C, Wiesenthal B, Dietz E, Ruden H. Survival of vancomycin-resistant and vancomycin-susceptible enterococci on dry surfaces. J Clin Microbiol 1998: 36: 3734–3736.
209. Siqueira JF Jr, Rôças IN. Campylobacter gracilis and Campylobacter rectus in primary endodontic infections. Int Endod J 2003: 36: 174–180.
210. Siqueira JF Jr, Rôças IN. Pseudoramibacter alactolyticus in primary endodontic infections. J Endod 2003: 29: 735–738.