| Oral Microbiol Immunol 1999: 14: 250–256 | Copyright © Munksgaard 1999 |
| Printed in Denmark · All rights reserved | Oral Microbiology and Immunology |
| ISSN 0902-0055 |
Molecular approaches to the differentiation of Actinomyces species
C. Jauh-Hsun, T. Vinh, J. K. Davies, D. Figdor
Department of Microbiology, Monash
University, Clayton, Victoria, Australia
Jauh-Hsun C, Vinh T, Davies JK, Figdor D. Molecular approaches to the differentiation of Actinomyces species.
Oral Microbiol Immunol 1999: 14: 250–256. C Munksgaard, 1999.
The definition of the genus Actinomyces relies heavily on traditional methods of taxonomy. This study sought to develop molecular tools for the identification of strains of Actinomyces israelii and Actinomyces gerencseriae. Oligonucleotide probes were designed and one of these successfully differentiated A. gerencseriae from ten strains of A. israelii and three other Actinomyces species by DNA:DNA hybridization. However, probes based on known 16S rRNA sequences failed to hybridize to all the strains previously identified as A. israelii. Using the PCR technique, a region encoding a portion of the 16S rRNA was amplified from genomic DNA. The results showed that A. israelii can be divided into three different groups based on comparison of the amplified DNA sequences. This information should allow the development of probes that are specific for these newly identified groups of strains within the species A. israelii.
Key words: Actinomyces israelii; Actinomyces gerencseriae; polymerase chain reaction; hybridization; DNA probe; identification; taxonomy
David Figdor, Medical Centre, 517 St Kilda Road, Melbourne, VIC 3004 Australia
Accepted for publication September 25, 1998
In human infections, Actinomyces israelii is the most common species causing actinomycosis (2, 27, 30). Typical symptoms of this chronic granulomatous disease include multiple abscesses, suppuration and draining sinus tracts (17). A. israelii has also been repeatedly identified in cases of failed endodontic therapy, where it causes a persistent extraradicular infection in the tissues, termed periapical actinomycosis (4, 12, 20, 33). The preferred treatment of periapical actinomycosis is surgical curettage, since the disease is resistant to conventional endodontic treatment and to routine administration of antibiotics. Clinical reports and our own laboratory studies show that, in the presence of an established actinomycosis, antibiotics are only likely to be effective when administered for periods of 6 weeks to several months (1, 24, 25).
Many scientific and clinical investigations have been hampered by problems in the identification of Actinomyces species. The sampling and laboratory growth of these organisms is challenging, so identification of Actinomyces species and the clinical diagnosis of actinomycotic infections are often based on histopathology of biopsy material. Such slow and complex procedures have led to clinical management that has been based on empirical treatment rather than on the true diagnosis of the presence of Actinomyces species. Therefore, efficient methods for the identification of A. israelii would be of considerable clinical value because such tools may lead to earlier and improved diagnosis of actinomycotic infections.
Conventional numerical taxonomic analyses have been used to define the genus Actinomyces (26). A. israelii is reasonably well separated from other Actinomyces species based on biochemical data (2, 14, 26, 31), and strains of A. israelii have been further characterized by serology into serotypes I and II (16, 26). However, these strains have since been reclassified (15) as two distinct species – A. israelii (formerly A. israelii serotype I) and Actinomyces gerencseriae (formerly A. israelii serotype II). Previous studies have reported that the species A. israelii and A. gerencseriae can be differentiated from each other by serological methods (10, 16, 31), polyacrylamide gel electrophoresis banding patterns (19), the ability to ferment arabinose (15) and 16S ribosomal RNA sequence data (32).
Data from 16S rRNA sequences are being used to provide an improved systematic structure based on an evolutionary relationship between organisms (21, 34, 35). These methods have been used to distinguish strains of Actinomyces species from other related genera and to separate Actinomyces species from each other in a phylogenetic tree (6, 22, 23, 32). Although the development of a comprehensive phylogenetic structure within the genus Actinomyces is still unfolding, genetic analysis of rRNA sequences has allowed the recognition of several new Actinomyces species (9, 23). The new species along with others in the genus are listed in Table 1.
In a first report of the 16S rRNA sequence of Actinomyces species (32), a species-specific oligonucleotide probe was designed for A. israelii. However, this probe did not allow differentiation of A. gerencseriae from A. israelii. In this report we describe the development of an oligonucleotide probe that is species-specific for A. gerencseriae and outline an attempt to clarify the phylogenetic position of some Actinomyces species using 16S rRNA sequence data.
Table 1. Human and animal Actinomyces species
|
Actinomyces bovis |
Actinomyces israelii |
|
Actinomyces denticolens |
Actinomyces meyeri |
|
Actinomyces europaeus |
Actinomyces naeslundii |
|
Actinomyces georgiae |
Actinomyces neuii subsp. anitratus |
|
Actinomyces gerencseriae |
Actinomyces neuii subsp. neuii |
|
Actinomyces graevenitzii |
Actinomyces odontolyticus |
|
Actinomyces hordeovulneris |
Actinomyces radingae |
|
Actinomyces howellii |
Actinomyces slackii |
|
Actinomyces humiferus |
Actinomyces turicensis |
|
Actinomyces hyovaginalis |
Actinomyces viscosus |
Material and methods
Bacteria and culture conditions
The Actinomyces species used in this study are shown in Table 2. The bacteria were cultured in brain heart infusion broth (1%, Oxoid, Basingstoke, UK) containing 0.02% (v/v) Tween 80, or on plates which were solidified by the addition of 1.5% (w/v) agar to the liquid media. Inoculated plates and broth were incubated at 37°C, anaerobically (10% CO2+10% H2 in N2) for 7–14 days.
Biochemical tests
All strains were checked using a biochemical test kit (Microbact 24AN System, Pacific Diagnostics) that consists of 23 substrates and a control. Strains were incubated at 37°C, for 7 days on brain heart infusion agar then suspended in broth by vortexing and 4 drops were dispensed into each test well in a microplate and overlaid with a drop of sterile mineral oil to prevent evaporation of test reagents. The microplate was incubated anaerobically at 95% humidity for 5 days and strains were identified according to the procedures in the Microbact 24AN System, which is based on the VPI anaerobe laboratory manual (13). The biochemical test results were analyzed by using a numerical taxonomy program (NTSYSPC). Tests were converted into 0 or 1, for a negative or positive reaction, respectively. The reference data for each species in the Microbact 24AN manual employed “w” (weak) and “v” (variable) values, which were also included into the character states (w=0.75, v=0.5).
DNA isolation
Bacteria were harvested, washed and suspended in 50 µl of 20 mg/ml lysozyme and 50 µl of 1 unit/µl mutanolysin, and incubated at 37°C for 30 min. Cells were disrupted by adding 5 µl of 20 mg/ml Proteinase K solution and 100 µl of Lysis Buffer (100 mM Tris-HCl pH 9, 20% sodium dodecyl sulfate [SDS], 25 mM EDTA, 300mM NaCl) at 56°C for 30 min with occasional inversion. Fifty µl of 5 M NaCl was added to the mixture before treating with an equivalent volume of chloroform (250 µl) to remove cell fragments. After separation of RNA and cell fragments by centrifugation, the supernatant was transferred to a new tube. DNA was precipitated with isopropanol and 4 µl of 3 M sodium acetate, pH 4.5 (20 min on ice), and the precipitate collected by centrifugation. The pellet of collected DNA was washed with 70% ethanol, dried in a Speedivac concentrator before being dissolved in 20 µl of sterile distilled H2O. DNA samples were separated by electrophoresis in a 1% agarose gel stained with ethidium bromide and run in a TAE buffer (0.04 M Tris-acetate, 0.001 M EDTA). DNA was visualized and photographed using ultraviolet light.
Table 2. Actinomyces strains used in this study
| Strain | Source | GenBank number | Strain history | Species identification |
| ATCC 121021 | 1 | AF058042 | Human brain abscess | A. israelii (formerly A. israelii serotype I) |
| ATCC 10048 | 1 | AF058046 | Human pleural fluid | A. israelii (formerly A. israelii serotype I) |
| L110B | 2 | AF058041 | Human dentine | A. israelii (formerly unknown serotype) |
| L104C2 | 2 | AF058044 | Human dentine | A. israelii (formerly unknown serotype) |
| L115B1 | 2 | AF058047 | Human dentine | A. israelii (formerly unknown serotype) |
| 12M | 2 | AF058043 | Human periodontitis | A. israelii (formerly unknown serotype) |
| 21Y | 2 | AF058045 | Human periodontitis | A. israelii (formerly unknown serotype) |
| 10AA | 2 | AF058048 | Human periodontitis | A. israelii (formerly unknown serotype) |
| AH | 3 | AF058040 | Human periapical lesion | A. israelii (formerly unknown serotype) |
| CCUG 35455 | 5 | AF058049 | Human infection | A. israelii |
| ATCC 121041 | 1 | AF058052 | Human sinus | A. naeslundii serotype I |
| ATCC 159871 | 1 | AF058051 | Hamster periodontal disease | A. viscosus serotype I |
| NCTC 9935 (= ATCC 179291) | 4 | AF058053 | Human deep caries | A. odontolyticus serotype I |
| CCUG 34703 (= ATCC 238601) | 5 | AF058050 | Human parotid abscess | A. gerencseriae (formerly A. israelii serotype II) |
1 Type strain
1. American Type Culture Collection, USA
2. Centre for Oral Health Sciences, Malmö, Sweden
3. Department of Endodontics, Umeå University, Sweden
4. National Collection of Type Cultures, UK
5. Culture collection, University of Göteborg, Sweden
Table 3. DNA sequences of each oligonucleotide used in this study
| Oligonucleotide | DNA Sequence | Description |
| Act300F | 5’ TGAGTAACACGTGAGTAACC 3’ | Forward primer |
| Act300R | 5’ AGAGGTTCACAACCCGAAGG 3’ | Reverse primer |
| Act-isr | 5’ CCAAAAACACCACAAAAGTG 3’ | Specific for A. israelii1 |
| Act-ger | 5’ CCAAAAACACCAAACAGTGC 3’ | Specific for A. gerencseriae |
1 Derived from Stackebrandt & Charfreitag (32)
Oligonucleotides
Oligonucleotides were synthesized on an Applied Biosystems 381A Oligonucleotide Synthesizer. The purification of oligonucleotides was carried out following the OPC purification protocol (PE Applied Biosystems, Melbourne). The DNA sequences of the oligonucleotides used in this study are shown in Table 3. Oligonucleotide probes were labeled by using the DIG Oligonucleotide 3’-end labeling Kit (Boehringer Mannheim, Germany) according to the manufacturer’s instructions.
DNA dot blots
Five µl of each genomic DNA sample (2 ng/µl) was dropped onto a positively charged nylon membrane, air dried and ultraviolet cross-linked for 5 min on a TFL-20M hybrid Crosslinker Transilluminator (Integrated Sciences, Melbourne). The membrane was then prehybridized in a sealed plastic bag with hybridization solution (5 × SSC [sodium chloride sodium citrate], 1% (w/v) blocking reagent, 0.1% (w/v) N-lauroylsarkosine, 0.02% (w/v) SDS) at 65°C for 5 h. After incubation, the hybridization solution was replaced with a hybridization solution containing probes (25 pg/ml) and then incubated at 48°C for oligonucleotide Act-isr, or 50°C for oligonucleotides Act300F and Act-ger. The membrane was removed from the plastic bag and washed for 5 min (2 times) at the same hybridization temperature with 2×SSC, 1% (w/v) SDS, 15 min (2 times) with 1×SSC, 1% (w/v) SDS, and 15 min (2 times) with 0.1×SSC, 1% (w/v) SDS. The detection of DNA probe by an enzyme-linked immunoassay (CDP-StarTM) was based on the procedure described in the Detection Kit (Boehringer Mannheim).
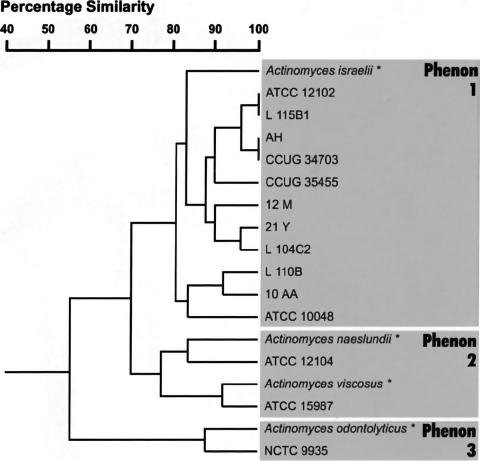
Fig. 1. Phenogram, based upon the simple-matching coefficient with clustering by the unweighted pair group method, showing similarity between Actinomyces strains using biochemical test data. The reference data from the Microbact 24AN manual are marked (*).
Amplification of DNA by PCR
Polymerase chain reactions (PCR) were performed in an FTS-1 Thermal Sequencer. The reaction mixture contained 10–50 ng/µl template DNA, 5.0 µM of each primer, 200 µM dNTPs (Promega), 0.5 µl Taq DNA polymerase (Boehringer Mannheim) and 10 µl 5× reaction buffer with 2.5 mM MgCl2, made up to a final volume of 50 µl with sterile distilled water. These components were mixed well and overlaid with 20 µl of paraffin oil. The PCR program consisted of 1 cycle of 95°C for 2 min, 50°C for 1 min, 72°C for 1 min, followed by 40 cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 1 min, and then a final extension at 72°C for 5 min, and a final hold at 4°C.
DNA sequencing and analysis
DNA sequencing was performed by the method outlined in the Prism Ready Reaction Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems) on an Applied Biosystems Model 373A DNA Sequencing System. Sequencing results were analyzed using the SequencherTM program (Gene Codes Corporation, Ann Arbor, MI). The primer used to sequence the PCR products was Primer Act300F. The DNA sequences were analyzed using a program, Pileup (www.gcg.com), which creates a multiple sequence alignment from a group of sequences using progressive pairwise alignments and plots a tree showing the clustering relationships used to create the alignment.
Results
Classification of Actinomyces strains based on biochemical test data
The phenetic resemblance for each strain was calculated in two ways. The first was the simple matching index, which computes various association coefficients for qualitative data, in which negative similarities are included. The second was by taxonomic distance, which computes various similarity or dissimilarity indices for interval measurement. The unweighted pair group method using arithmetic averages was employed for clustering. Both methods resulted in similar phenograms; the simple matching index is shown in Fig. 1. All A. israelii and A. gerencseriae strains, including type strains, were grouped in phenon 1 at similarity levels of 80% to 100%. It should be noted that only A. israelii ATCC 10048 appeared to ferment arabinose in these tests. The second phenon contained two distinct sub-clusters, Actinomyces naeslundii and Actinomyces viscosus, which were joined together at the 77% similarity level. The third phenon contained the strain NCTC 9935, linked to the reference data for Actinomyces odontolyticus at the 87% similarity level.
DNA dot blots
Genomic DNA from all Actinomyces species was hybridized with oligonucleotide probes Act300F, Act-isr and Act-ger (Fig. 2). The Act300F probe used in this experiment acted as a positive control since it contained a sequence that was highly conserved in all Actinomyces species. The Act300F probe hybridized strongly to genomic DNA from all Actinomyces species tested (Fig. 2, top panel). The Act-isr oligonucleotide probe (Table 3) has been reported to be specific for A. israelii serotype I strains (32). This Act-isr probe only hybridized intensely to DNA from strains AH and ATCC 12102, and weakly to DNA from strain L110B (Fig. 2, middle panel). The Actisr probe therefore failed to hybridize to the genomic DNA from the majority of the strains previously identified as A. israelii. A specific probe for A. gerencseriae, Act-ger, was designed based on previously published sequencing data (32). The Act-ger probe (Table 3) was derived from a divergent sequence for A. gerencseriae, at the position used for the Act-isr probe for A. israelii (serotype I). Using the Act-ger probe, a positive hybridization signal was obtained only from the strain CCUG 34703, the type strain for A. gerencseriae (Fig. 2, bottom panel).
Analysis of genomic DNA encoding part of 16S rRNA of Actinomyces species
The hybridization data described above suggested that there might be greater 16S rRNA sequence diversity amongst these strains than had previously been suspected. The oligonucleotide primers Act300F and Act300R were therefore designed based on the partial 16S rRNA sequences of Actinomyces species. These oligonucleotides were used to amplify, by PCR, a portion of the region previously studied (32). PCR products were separated by agarose gel electrophoresis, and the desired fragments were purified and sequenced using oligonucleotide Act300F as a primer. The sequenced region had a consensus length of 291 nucleotides, including a region that was not previously sequenced. The GenBank accession numbers are listed in Table 2.
The alignment between these sequences and those already published indicates that Actinomyces species have a high degree of sequence identity. It is also evident that the region described as highly variable (32) is more complex than previously suspected. The sequences found in A. israelii strains ATCC 10048, L115B1 and CCUG 35455 were the same as that reported before (A. israelii strain DSM 43020, GenBank accession number X53228) (32), however the other six strains varied in this region.
DNA sequence identity between each strain was calculated and used to construct a diagram showing the similarity between the determined sequences (Fig. 4). Based on the partial 16S rRNA sequences, the strains of A. israelii were divided into three groups (Fig. 4).
Group 1. The sequence of the region amplified from strains L115B1 and CCUG 35455 was identical, or almost identical, to reference strain ATCC 10048. The strain 10AA was located outside of the cluster, but had a slightly higher DNA homology (93%) to strain ATCC 10048. This group of A. israelii strains has a high similarity (97%) with Propionibacterium acnes based on the partial 16S rRNA sequence (5).
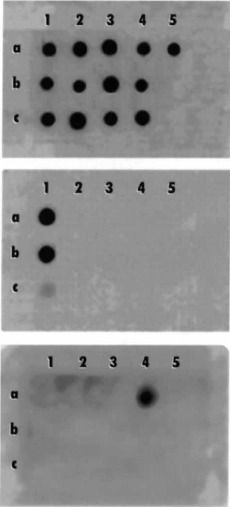
Fig. 2. DNA dot blotting using three oligonucleotide probes that were hybridized with genomic DNA from various Actinomyces strains. The DNA preparations 1a, b and c are from A. israelii strains AH, ATCC12102 and L110B; dot blots 2a, b and c are A. israelii strains 12M, L104C2 and 21Y; dot blots 3a, b and c are A. israelii strains ATCC 10048, L115B1 and 10AA; dot blots 4a, b and c are A. gerencseriae CCUG 34703, A. naeslundii ATCC 12104 and A. viscosus ATCC 15987 and dot blot 5a is A. odontolyticus NCTC 9935.
Top panel shows a positive hybridization signal to all Actinomyces species, using the Act 300F probe. Middle panel shows hybridization using the Act-isr probe, which is positive for A. israelii strains AH and ATCC 12102, and weakly positive for strain L110B. Bottom panel shows hybridization with the Actger probe, which is positive only for A. gerencseriae CCUG 34703.
Group 2. This cluster contained three A. israelii strains, 21Y, L104C2 and 12M, which had a close similarity in DNA sequence with each other (94–97%), but had a lower DNA homology with the two A. israelii reference strains ATCC 12102 and ATCC 10048 (80–85%).
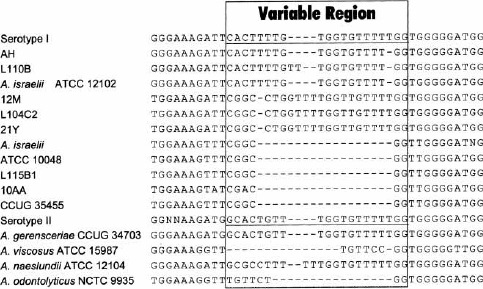
Fig. 3. Alignments of genomic DNA sequences encoding part of the 16S rRNA. The sequences labeled serotype I and serotype II are those described by Stackebrandt & Charfreitag (32); the sequence labeled A. israelii is that from strain DSM 43020 of A. israelii serotype I (GenBank accession number X53228). The sequences used to design oligonucleotides Act-isr and Actger are underlined in the serotype I and serotype II sequences, respectively. The box delineates the region of variable sequence, described previously (32).
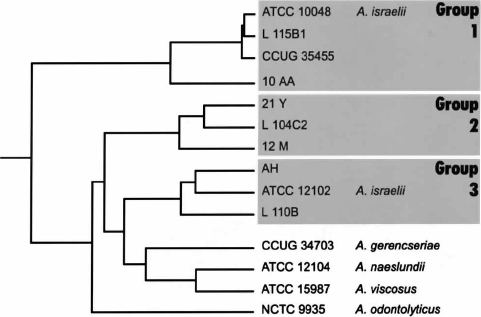
Fig. 4. Diagram, based on progressive pairwise alignment analysis, showing the relationship between genomic DNA sequences encoding part of the 16S rRNA in various Actinomyces strains.
Group 3. This cluster contained strains ATCC 12102, AH and L110B. Other than a minor disparity in a few positions, sequences of strains AH (95%) and L110B (94%) were very similar to the type strain, A. israelii (ATCC 12102).
The A. gerencseriae strain CCUG 34703 was grouped with A. naeslundii (ATCC 12104) and A. viscosus (ATCC 15987). The A. gerencseriae strain CCUG 34703 showed a lower DNA homology (82–88%) with the three groups of A. israelii as compared to A. naeslundii (92%) and A. viscosus (91%), which confirms an earlier report (32). The sequences from other Actinomyces species (A. naeslundii, A. viscosus and A. odontolyticus) were identical to published 16S rRNA sequences.
Discussion
The identification of Actinomyces species has been repeatedly described as a challenging problem (10, 15, 22, 31), yet reliable and rapid differentiation tools would be of considerable value for the clinical diagnosis and management of actinomycosis. Apart from traditional methods of colony morphology, Gram stain and biochemical tests (2, 3, 30, 31), other tools available for classification have included serology (10, 11, 16, 18) and SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of whole cell proteins (19). Recently, molecular techniques have rejuvenated the approach to the problems of identification and taxonomy and have been applied to distinguish Actinomyces species (9, 22, 23). Here, we describe for the first time the successful development and testing of an oligonucleotide probe that is species-specific for A. gerencseriae. In addition, 16S rRNA from a number of A. israelii strains was sequenced and the results showed that there is considerably more heterogeneity than was previously believed.
The identities of all strains used in this study were initially confirmed using traditional biochemical tests for classification of Actinomyces strains. When the strains were grouped together with reference strains in a phenogram (Fig. 1), the groups matched the patterns described for Actinomyces species in earlier studies based on numerical taxonomy (26, 28). All strains of A. israelii and A. gerencseriae were grouped together within a single phenon. The results from these biochemical tests did not allow differentiation between A. israelii and A. gerencseriae. Apart from one A. israelii strain (ATCC 10048), the other A. israelii strains failed to ferment arabinose, despite the suggestion that the majority of strains of A. israelii (89%) can be differentiated by their ability to ferment arabinose (15). Other studies have reported a lower proportion of strains that ferment arabinose (31) and that the fermentation of arabinose by A. israelii can be influenced by the basal medium (30). These results, therefore, confirm earlier reports of the difficulty of differentiation between A. israelii and A. gerencseriae based solely on biochemical tests.
Using a highly variable region of the partial 16S rRNA sequence described previously (32), several oligonucleotide probes were designed (Table 3). The Act-isr probe, which was identical to the A. israelii–specific oligonucleotide described by Stackebrandt & Charfreitag (32), failed to hybridize to genomic DNA samples of many A. israelii strains. This suggests that this probe cannot be used with the certainty that it will successfully detect all A. israelii strains in a given sample. The Act-ger probe was demonstrated to specifically hybridize to the genomic DNA isolated from the A. gerencseriae strain CCUG 34703 (Fig. 2, bottom panel). The deployment of the Act-ger probe should allow the detection of A. gerencseriae in a sample and it can be used to differentiate A. gerencseriae from A. israelii and other Actinomyces species.
The most comprehensive study to date of 16S rRNA sequences of A. israelii and A. gerencseriae has been that of Stackebrandt and Charfreitag (32). The data presented here suggests that the taxonomic position of the species A. israelii is more complicated than outlined in earlier reports. Three groups of sequences were found in A. israelii strains, one of which matched the sequence reported earlier (32) (Fig. 4, group 3), and two additional, distinct groups of DNA sequences that have not been described previously. The group 1 sequence (Fig. 4) is closely related to that from P. acnes and shows only slightly higher DNA homology to other A. israelii groups. However, A. israelii and P. acnes strains can be easily distinguished by biochemical tests (14, 29). The second additional sequence (group 2, Fig. 4) is more closely related to A. naeslundii (89%) than the other A. israelii groups (85% and 80%). Because there is no additional information regarding this group, it cannot currently be determined whether this group is a member of A. israelii, A. naeslundii or a new Actinomyces species. It should be noted that an earlier report, using an indirect immunofluorescence assay (29), found that A. israelii (serotype I) had three sub-groups, in addition to A. gerencseriae. Although it has not been tested, it is possible that the three sub-species described in serological data (26, 29) could correspond to the three A. israelii groups defined by 16S rRNA data in this study.
The third group (Fig. 4), which contained the type-strain ATCC 12102, included a strain that is known to be pathogenic. This isolate, strain AH, was recovered from a failed case of endodontic therapy (33) and has been demonstrated to be capable of inducing an experimental actinomycosis in animals (8). This raises the question as to whether this group of A. israelii contains bacteria that might represent a more virulent strain or species. Although it is not currently known, this issue could be resolved by comparing strains in an in vivo model similar to that used previously (8). The pathogenicity of the third strain (L110B) in this group has not been tested; however, strain L110B is known to have a cell surface ultrastructure that differs from strain AH (7). A recent review of Actinomycete infections revealed that the highest proportion of isolates from typical human actinomycotic abscesses were A. israelii species (about 56%) but that almost 25% of isolates were A. gerencseriae (27), which denotes the clinical importance of suitable differentiation procedures. Further tests in an in vivo model would be valuable in clarifying the relative pathogenicity of A. gerencseriae compared with strains of A. israelii.
In summary, the molecular methods described here have provided a new means for the differentiation and identification of Actinomyces species. An oligonucleotide probe has been developed that is species-specific for A. gerencseriae, and using this probe, it is possible to readily differentiate A. gerencseriae from A. israelii. The results also suggest that the taxonomic situation within the species A. israelii is more complicated than previously suspected. The sequence data generated in this study should also allow the differentiation by similar means of the three groups identified within A. israelii. These oligonucleotides could form the basis for further specific, rapid tests for the identification of strains responsible for actinomycotic infections.
Acknowledgments
We thank G. Sundqvist, Umeå University, S. Edwardsson, Malmö and E. Falsen University of Göteborg for supplying bacteria and G. Sundqvist, Sweden for constructive criticism. This study was supported by grants from the Australian Dental Research Fund Inc. and the Monash University Research Fund.
References
1. Barnard D, Davies J, Figdor D. Susceptibility of Actinomyces israelii to antibiotics, sodium hypochlorite and calcium hydroxide. Int Endod J 1996: 29: 320–326.
2. Borssén E, Sundqvist G. Actinomyces of infected dental root canals. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1981: 51: 643–648.
3. Brander MA, Jousimies-Somer HR. Evaluation of the RapID ANA II and API ZYM systems for identification of Actinomyces species from clinical specimens. J Clin Microbiol 1992: 30: 3112–3116.
4. Byström A, Happonen R-P, Sjögren U, Sundqvist G. Healing of periapical lesions of pulpless teeth after endodontic treatment with controlled asepsis. Endod Dent Traumatol 1987: 3: 58–63.
5. Charfreitag O, Stackebrandt E. Inter- and intrageneric relationships of the genus Propionibacterium as determined by 16S rRNA sequences. J Gen Microbiol 1989: 135: 2065–2070.
6. Embley TM, Stackebrandt E. The molecular phylogeny and systematics of the Actinomyces. Annu Rev Microbiol 1994: 48: 257–289.
7. Figdor D, Davies J. Cell surface structures of Actinomyces israelii. Aust Dent J 1997: 42: 125–128.
8. Figdor D, Sjögren U, Sörlin S, Sundqvist G, Nair PNR. Pathogenicity of Actinomyces israelii and Arachnia propionica: experimental infection in guinea pigs and phagocytosis and intracellular killing by human polymorphonuclear leukocytes in vitro. Oral Microbiol Immunol 1992: 7: 129–136.
9. Funke G, Alvarez N, Pascual C, Falsen E, Åkervall E, Sabbe L, Schouls L, Weiss N, Collins MD. Actinomyces europaeus sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol 1997: 47: 687–692.
10. Gerenscer MA, Slack JM. Serological identification of Actinomyces using fluorescent antibody techniques. J Dent Res 1976: 55: A184-A191.
11. Gohean RJ, Pantera EA, Schuster GS. Indirect immunofluorescence microscopy for the identification of Actinomyces sp. in endodontic disease. J Endod 1990: 16: 318–322.
12. Happonen R-P. Periapical actinomycosis: a follow-up study of 16 surgically treated cases. Endod Dent Traumatol 1986: 2: 205–209.
13. Holdeman LV, Cato EP, Moore WE. Anaerobe laboratory manual. 4th edn. Blacksburg, VA: VPI Anaerobe Laboratory, Virginia Polytechnic Institute and State University, 1977.
14. Holmberg K, Nord CE. Numerical taxonomy and laboratory identification of Actinomyces and Arachnia and some related bacteria. J Gen Microbiol 1975: 91: 17–44.
15. Johnson JL, Moore LVH, Kaneko B, Moore, WEC. Actinomyces georgiae sp. nov., Actinomyces gerencseriae sp. nov., designation of two genospecies of Actinomyces naeslundii, and inclusion of A. naeslundii serotypes II and III and Actinomyces viscosus serotype II in A. naeslundii genospecies 2. Int J Syst Bacteriol 1990: 40: 273–286.
16. Lambert FW, Brown JM, Georg LK. Identification of Actinomyces israelii and Actinomyces naeslundii by fluorescent-antibody and agar-gel diffusion techniques. J Bacteriol 1967: 94: 1287–1295.
17. Lerner PI. The lumpy jaw. Cervicofacial actinomycosis. Infect Dis Clin North Am 1988: 2: 203–220.
18. Leslie DE, Garland SM. Comparison of immunofluorescence and culture for the detection of Actinomyces israelii in wearers of intra-uterine contraceptive devices. J Med Microbiol 1991: 35: 224–228.
19. McCormick SS, Mengoli HF, Gerencser MA. Polyacrylamide gel electrophoresis of whole-cell preparations of Actinomyces spp. Int J Syst Bacteriol 1985: 35: 429–433.
20. O’Grady JF, Reade PC. Periapical actinomycosis involving Actinomyces israelii. J Endod 1988: 14: 147–149.
21. Olsen GJ, Woese CR. Ribosomal RNA: a key to phylogeny. FASEB J 1993: 7: 113–123.
22. Pascual Ramos C, Foster G, Collins MD. Phylogenetic analysis of the genus Actinomyces based on 16S rRNA gene sequences: description of Arcanobacterium phocae sp. nov., Arcanobacterium bernardiae comb. nov., and Arcanobacterium pyogenes comb. nov. Int J Syst Bacteriol 1997: 47: 46–53.
23. Pascual Ramos C, Falsen E, Alvarez N, Åkervall E, Sjödén B, Collins MD. Actinomyces graevenitzii sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol 1997: 47: 885–888.
24. Peabody Jr JW, Seabury JH. Actinomycosis and nocardiosis. A review of basic differences in therapy. Am J Med 1960: 28: 99–115.
25. Rippon JW. Actinomycosis. In: Rippon JW, ed. Medical mycology. 3rd edn. Philadelphia: Saunders, 1988: 30–53.
26. Schaal KP. Genus Actinomyces. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG, ed. Bergey’s manual of systematic bacteriology, vol. 2. Baltimore: Williams & Wilkins, 1986: 1383–1418.
27. Schaal KP, Lee H-J. Actinomycete infections in humans – a review. Gene 1992: 115: 201–211.
28. Schaal KP, Schofield GM. Current ideas on the taxonomic status of the Actinomycetaceae. In: Schaal KP, Pulverer G, ed. Actinomycetes. Zbl. Bakt. Suppl 11. Stuttgart: Gustav Fischer Verlag, 1981: 67–78.
29. Schaal KP, Schofield GM. Classification and identification of clinically significant Actinomycetaceae. In: Biological, biochemical, and biomedical aspects of Actinomycetes. London: Academic Press, 1984: 505–520.
30. Slack JM, Gerencser MA. Actinomyces, filamentous bacteria. Biology and pathogenicity. Minneapolis: Burgess Publishing, 1975.
31. Slack JM, Landfried S, Gerenscer MA. Morphological, biochemical, and serological studies on 64 strains of Actinomyces israelii. J Bacteriol 1969: 97: 873–884.
32. Stackebrandt E, Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii–specific oligonucleotide probe. J Gen Microbiol 1990: 136: 37–43.
33. Sundqvist G, Reuterving C-O. Isolation of Actinomyces israelii from periapical lesion. J Endod 1980: 6: 602–606.
34. Tanner A, Maiden MFJ, Paster BJ, Dewhirst FE. The impact of 16S ribosomal RNA-based phylogeny on the taxonomy of oral bacteria. Periodontol 2000 1994: 5: 26–51.
35. Woese CR. Bacterial evolution. Microbiol Rev 1987: 51: 221–271.