Endodontic Topics
| Endodontic Topics 2011, 18, 62–77 | 2011 © John Wiley & Sons A/S |
| All rights reserved | ENDODONTIC TOPICS 2011 1601-1538 |
Survival against the odds: microbiology of root canals associated with post-treatment disease
DAVID FIGDOR & KISHOR GULABIVALA
Most periapical lesions resolve after root canal treatment of teeth with primary infections. Over the last decade there has been a renewed focus on post-treatment apical periodontitis and its etiology. This review describes the microbiota associated with persistent post-treatment infection, including microbial identification, ecology, and environmental selection. Compared with untreated teeth, the infection pattern in root canals with post-treatment disease shifts to a resistant, mainly Gram-positive community. The main challenge in root canal re-treatment is access to the residual apical infection. Elimination of the microbial flora, or a dramatic reduction and sufficient ecological shift to allow host tissue healing, remains the biological goal.
Received 28 January 2011; accepted 4 February 2011.
Over millions of years, oral micro-organisms have evolved to co-exist with the mammalian host (1) mainly as biofilms adherent to enamel, dentin, cementum, and the oral mucosa. A dramatic shift in the biofilm ecology is essentially responsible for the development of caries and periodontal disease (2). In contrast, the dental pulp is an intrinsically sterile tissue and apical periodontitis occurs after a breach of the protective enamel/dentin encasement and polymicrobial invasion and infection of the pulp space.
Apical periodontitis is a more widespread disease than moderate or severe marginal periodontitis, affecting 50% of the population by age 50, and 62% of individuals over age 60 (3). Since the prime etiology is infection, clinical management of apical periodontitis is targeted at microbial control by root canal treatment. Many millions of root canal treatments are performed annually (4) and it is estimated that there are many hundreds of millions of root-filled teeth in the adult populations of Western countries (5).
The delivery of sound treatment with successful outcomes is a primary clinical goal. It is natural to expect a higher success rate for root canal treatment when it is performed by trained specialists rather than general practitioners. Based on epidemiological data, the success rate of root canal treatment performed in specialist practice is in the order of 87% compared with 72% for treatment in general practice (3). Although seemingly insignificant, the 15% difference equates to many millions of failed treatments when applied to Western populations (5). In the context of the cost of root canal re-treatment and crown or restoration replacement, the cumulative economic impact is in the order of billions of dollars. Thus, there are broad health, social, and economic consequences of root canal treatment failure and potentially significant benefits individually and collectively if it were possible to reduce the proportion of endodontic failures (5).
Different types of infection
The prime reason for post-treatment disease is infection in the apical part of the root canal system by species that have endured or evaded antimicrobial treatment, survive in the filled root canal, and are capable of inflaming the periapical tissue. Some species are well suited to the barren but protected environment of the filled root canal, yet how microbes take advantage of this unique situation has not been adequately elucidated at this time.
There are two potential pathways for post-treatment infection (Fig. 1). The first is for micro-organisms that were part of the initial polymicrobial consortium of the untreated root canal to prevail and subsequently participate in post-treatment disease. An ability to endure biomechanical preparation is invaluable, but alone is not enough without a capacity to survive in the root-filled canal and inflame the periapical tissue. A second pathway is re-infection of the root-filled canal via microbial leakage around the coronal restoration. It should be noted that this conceptual framework is deduced from currently available information on the composition of the microbial flora in untreated and treated canals, and that direct evidence is lacking to illustrate the natural pathogenesis of the respective infection patterns.
Species that survive treatment
A few studies have provided information on the species that survive instrumentation and antimicrobial irrigation. Generally, Gram-negative species and fastidious anaerobes have a limited capacity to endure treatment and are readily eliminated by instrumentation and antimicrobial irrigation (6). Gram-positive species and facultative anaerobes have a higher rate of recovery in post-instrumentation samples (6–12). It is worth noting that after root canal preparation there are low cell numbers that survive treatment, in contrast to the high initial microbial cell population (7, 13, 14). Any cells surviving treatment are likely to do so in small biofilms and discrete pockets of cells away from the main body of the canal (15, 16). Since there are few surviving cells that are in a vulnerable state, there are implications for sample collection because special methods are necessary for recovery by culture (17). Without these measures, it is highly likely that the sampling procedure will not pick up surviving cells.
Species that survive in the root-filled canal and maintain periapical inflammation
Although some species may elude antimicrobial treatment, if they are left without suitable nutrition and are not in a propitious location, the microbes may not be able to survive in the root canal over time. A number of microbial properties favor the selection and survival of some species over others. For example, an ability to endure starvation is a beneficial survival characteristic which helps ensure that some species outlast others until they may access nutrients from the local milieu, e.g. serum-type fluid that may seep into the canal space over time.
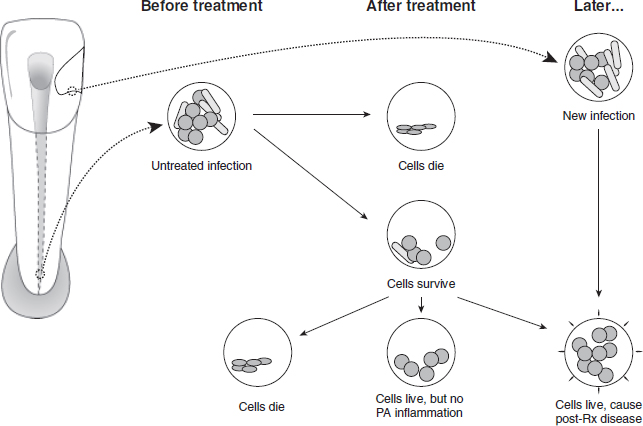
Fig. 1. Potential pathways for post-treatment infection.
Starvation survival characteristics have been demonstrated in selected species known to be involved in persistent infection such as Enterococcus faecalis (18) and Candida albicans (19), whereas others seldom identified in persistent infection have poor starvation survival ability (20).
Although some microbes may weather treatment, their subsequent destiny and involvement in maintenance of the periapical lesion depends on nutritional availability, luck, and location (21). For example, microbes that survive in the isthmus between canals in the mesial root of a lower molar after endodontic treatment (16) may not have the ability to communicate with the periapical tissue and thus not be in a position to inflame host tissue.
Re-infection after treatment
Sound restoration goes hand-in-hand with high quality root canal treatment for an optimal outcome of the treated tooth. Any breach in the marginal integrity of the coronal restoration poses a threat of nutrient or microbial leakage into the root canal system and an ensuing risk to the prognosis. Based on cross-sectional studies, it has been observed that there is a higher incidence of periapical pathoses when there are radiographic signs of poor quality restorations (22–24). While it makes good biological sense that there is a connection between coronal microleakage and the outcome of root canal treatment, some studies point to the root filling as an effective barrier to apical microbial advancement (25). It remains to be shown whether the relationship between poor coronal seal and reduced treatment outcome is a reflection of the (lower) quality of the initial treatment or that coronal leakage was responsible for the subsequent treatment outcome.
Specific versus non-specific microbial etiology
The seminal discovery and publication of Koch’s work on anthrax (26) led to the realization that many classic diseases are the result of individual pathogens with a specific microbial etiology. Yet the insightful microscope and culture observations of Miller in 1894 (27) showed that apical periodontitis is associated with a collection of species in a polymicrobial infection. That the infection is polymicrobial means that it is difficult to attribute specific roles to individual species. Thus, all species present and living have the potential to contribute to the infection process (21) and, together, they may act collectively for the benefit of the community (28). Factors such as strain variation may also account for variable pathogenicity (29–31).
The fundamental reason for accurate and complete identification of bacterial communities from root canal systems is to disclose those species or species-combinations that may play key roles in the progress of the disease or its acute exacerbation, especially those that may be resistant to conventional therapy or implicated in treatment failure (32, 33).
The association between the type of root canal flora or individual species and periapical lesion development has been studied by several groups of investigators. Classic studies on the periapical responses to indigenous bacterial infections in the monkey model used an experimental design in line with Koch’s postulates for a polymicrobial infection (34–36). Teeth that were infected by exposure to the oral cavity developed periapical lesions; the proportion of facultative anaerobic species decreased and strict anaerobic species increased over a 6–7 month period (36). There are changes in the type and proportions of the flora over time as well as location within the root canal, as shown by animal experiments in monkey (35), dog (37), and rat teeth (38), which induce species-specific host responses (39, 40).
Species work together to support growth and survival and co-operate to thwart the host defense. Other strategies may include gene transfer within the biofilm matrix for the purpose of enhancing virulence or evading the host response (41, 42). Periapical lesion development therefore seems to be dependent upon the nature of the mixed infection, the succession within it, and its ultimate survival. Some species, however, are capable of survival and host stimulation as sole pathogens (34, 43).
The clinical spectrum of apical periodontitis varies from acute to chronic with suppurative presentations and the range of responses may be a function of variation in host response (44) and/or a function of fundamentally different types of microbial flora (40). Different pathogenic profiles within the microbiota raise the prospect of different susceptibilities to treatment and the prospect of specific treatment protocols.
Biofilm ecology
Medical microbiology has been founded on treating infections conceptualized as individual planktonic cells colonizing tissue surfaces and compartments. It has become apparent that this perception of independent single cells, which survive by moving towards nutrients and away from harm, is actually not the preferred mode of bacterial survival. The natural pattern is a sedentary existence in enclosed extracellular matrix communities that are frequently multi-species in composition and adherent to surfaces. This is a basic description of biofilms, which were first recognized in root canal systems by Nair (45). Although the term “biofilm” was not used, Nair (45) described the presence of clusters of cells thus:
Most of the flora in the root apex remained suspended in an apparently moist canal lumen. Less frequently, dense aggregates of bacteria could be observed sticking to the dentinal wall of the root canal. The interbacterial spaces revealed an amorphous material, particularly towards the center of the aggregates. The single or multi-layered bacterial condensations were visible only at EM level.
Subsequently, he revised this interpretation to suggest that the majority of the bacteria were probably embedded in a surface adherent biofilm (personal communication), a view confirmed in other studies (46). The study of endodontic biofilms is in its infancy, but it is fortunate that the process of biofilm formation follows a universal pattern and Endodontics can borrow many established concepts (47).
The survival attributes of a motile single cell are dramatically different from those required for sedentary community life. Precisely what surface conditions encourage colonization on root canal dentin remains to be determined (48), but colonizing species act together to optimize the utilization of local resources (43, 49). Such sophisticated bacterial collaboration requires intercellular communication. Cells release specific signal molecules into the environment and, through diffusion or convective distribution and detection by other cells, they communicate when they have reached a critical cell density (50–52). In this way, the collective population co-ordinates efforts in nutritional utilization, virulence expression, and various other community functions. Thus, the extracellular matrix develops as a co-ordinated affair, allowing fluids to circulate and an exchange of cellular components and nutrients amongst the microbial community (53, 54).
Some cells play an important role in the survival of the community. Persister cells neither grow nor die (55) and are thought to have some responsibility for resistance to antimicrobials (56). Biofilms may also enhance their chances of survival by dispersing breakaway portions or individual cells in a planktonic phenotype (57). It is therefore possible to visualize localized variations in biofilm composition in the root canal system, for which some evidence exists (46, 58, 59).
In summary, biofilms are highly organized multicellular, multi-species structures that collaborate collectively to harvest nutrients, and display sensing and survival tactics. There is a need for more sophisticated strategies to control biofilms, not only in chemomechanical terms but especially in biological terms (53, 60). In the untreated infected root canal, there is a complex and rich biofilm community, but biomechanical instrumentation results in a dramatic change to the environment. Root canal treatment causes massive disruption and dissolution of the biofilm and strips away most of the nutrient supply. In cases treated in accordance with accepted principles, only small pockets of bacterial communities may survive in limited spaces of the root-filled canal (16). How long they survive after root filling and whether they evoke post-treatment disease is currently unknown.
Microbiology of untreated root canal infection
The hard and soft tissues of the oral cavity are home to a rich and diverse assortment of micro-organisms, previously estimated with culture methods at >4500 different kinds (61), and with molecular techniques and re-classification this number has more recently been revised upwards to >700 species (62). Each individual is estimated to have 100–200 species of oral bacteria (63) and all have the potential to invade the pulp space, yet a relatively small number of species are typically recovered in root canal samples. Using advanced culture techniques, a limited assortment of species has been reported (33, 64–66), typically a polymicrobial mix of 2–12 species.
Molecular methods have the potential to recover DNA from difficult-to-culture species and, while a broader range of species-specific DNA has been described (28, 67, 68), the number and diversity of species in individual root canals is typically 10–30 species (28, 68), which remains well below that reported in the oral cavity (69) or identified in the periodontal pocket (63).
Strong selection pressures define the type and course of infection. The local environment primarily governs microbial composition and organization, and the most important factors are anaerobiosis, microbial interactions and nutrition. Obligate anaerobes dominate the microbial flora in root canal infections of human teeth with intact pulp chambers (33, 64–66). Several studies have shown that particular species have a tendency to associate together (70–72), and there is good reason for microbes to co-operate collectively so that they can flourish in a communal environment. Individual species may supply essential nutrients for the growth of other members of the population (73–75). Bacteria may also compete for nutrition, or inhibit the growth of others by production of bacteriocins (76).
Nutrition is essential for microbial growth – nutrients may potentially be derived from the oral cavity, connective tissue and blood components in degenerating pulp (77), dentinal tubule contents, or a serum-like fluid from periapical tissue (78).
Thus, apical periodontitis is caused by a habitat-adapted polymicrobial infection of the pulp space. The microbial flora typically consist of a restricted group of species, with equal proportions of Gram-positive and Gram-negative species, dominated by anaerobes with fastidious environmental and nutritional requirements (33, 75).
Ecology and species
The root canal infection is a dynamic process and various bacterial species dominate at different stages of this process. In a long-standing infection, there is a shift towards dominance of the community by selected species. The most important factors driving this development are availability of nutrition, oxygen level (redox potential), and the local pH within the root canal (21, 33).
Exogenous nutrients, such as fermentable carbohydrates, can affect the microbial ecology of the coronal parts of an exposed root canal, but endogenous proteins and glycoproteins are the principal nutrients in the main body of the root canal system (21, 79, 80). Although there is a restricted supply of proteins in the root canal from progressive degradation of the small volume of pulpal tissue, the bacteria induce a periapical inflammation that leads to influx of a serum-like exudate into the canal. This fluid is a sustainable nutrient source containing proteins and glycoproteins for those bacteria that have a proteolytic capacity. The bacteria which dominate this stage of the infection are likely to be those that either have proteolytic capability, or maintain a co-operative synergy with those that can utilize this substrate for bacterial metabolism (21, 79, 80). Bacterial metabolism of the serum-like fluid also causes reduction of the redox potential and a concomitant rise in the pH (2).
Differences between culture and molecular results
Elaboration of the root canal microbial flora was established using sophisticated anaerobic culture methods that were specifically developed for growing many obligately anaerobic human pathogens (81). Advances in molecular techniques have increased our ability to differentiate bacteria and led to the establishment of new genera and species. Many of the taxonomic refinements have developed as split-offs from previously established genera and species. Because classical culture and molecular methods both have distinct advantages and drawbacks, it makes good sense to recognize their respective attributes in order to gain the most from studies on root canal microflora. Successful cultivation relies on viable micro-organisms that live and grow on plates or in liquid media. Molecular approaches, specifically PCR, depend on isolation and amplification of target-specific DNA, regardless of cell viability. With both approaches, the sample is obtained in the same way by soaking fluid from the root canal onto paper points and storing in transport medium. Thereafter, processing differs in the laboratory. Some of the respective features are summarized in Tables 1 and 2.
Conventional bacterial identification has been based on an array of biochemical tests, which are particularly technique- and operator-sensitive. Relatively minor differences in culture procedures can influence results and some characteristics may not be reproducible under conditions used in different laboratories. In addition, the biochemical tests are greatly influenced by the size and age of the inoculum and the degree of anaerobiosis (33). In early studies, isolates were identified only to the genus level. By 1974, the first thorough bacteriological analysis to the species level was published (64), which was subsequently followed by others who adopted more comprehensive procedures for identification (65, 66, 82).
Bergey’s Manual of Systematic Bacteriology (83) was used as the early standard for identification (32, 81, 84, 85). Later, others (65, 66, 82) adopted the VPI manual (86) for anaerobic bacteria. The introduction of biochemical test kits simplified laboratory procedures (87) and from the 1980s the API kits became popular (88). These offered a degree of standardization across laboratories (89) and were often supplemented with other tests (Bergey’s and the VPI manual) as their databases were unable to identify all endodontic isolates (89–92). More recently, commercially available kits have been used exclusively with identification based entirely on the kit database (93–97). This is problematic because the databases do not cover the strains found in root canal systems (98).
Table 1. Advantages and disadvantages of culture-based methods
| Advantages | Disadvantages |
| Assesses living (culturable) microbes | Risk of contamination |
| Able to recognize viable cells in a sample | High skill level is necessary for optimal results |
| Easy to quantitate cells in a sample | Time and resource intensive |
| High sensitivity with appropriate media | Relies on phenotypic biochemical characterization |
The use of molecular techniques for identification of bacteria in Endodontics is relatively recent (99). The studies have used the comparative 16S rRNA gene sequence approach to identify bacteria in root canal samples. The majority have applied species-specific PCR amplification of variable 16S rRNA sequences for identification. Some of the target species include Actinomecetales (99), Fusobacterium (99–102), Bacteroides forsythus (99–101, 103–105), Streptococcus species (99, 102, 104), Prevotella intermedia and nigrescens and black pigmented species (106, 107), Treponema (105, 108–110), Slackia exigua, Mogibacterium timidum and Eubacterium saphenum (111), Prevotella spp. (101), Porphyromonas spp. (101, 105, 109), Peptostreptococcus spp. (101), and Enterococcus faecalis (101, 112).
Some studies have used DNA hybridization probes to detect specific bacteria in samples (100, 103, 104, 113). Few of these studies have conducted sensitivity/specificity checks on their DNA probes (111) and few have sequenced amplicons to confirm identity (67, 68, 99, 109, 111, 112, 114). Only a few studies (67, 68) attempted to carry out a microbial community analysis using both cultivation and molecular cloning approaches.
The influence of various factors in sampling, detection, and identification method on species recovery was shown in a study that combined both methods (115). A clinically intact tooth with a periapical radiolucency, extracted for restorative reasons, was immediately placed in an anaerobic chamber and surface-decontaminated; the pulp chamber was accessed and the root canal was conventionally sampled with paper points. The tooth was then cryo-pulverized for the second sample. Both samples were analyzed by culture-dependent and culture-independent molecular techniques (16S rDNA amplification, cloning, sequencing). Samples of the tooth yielded 44 taxa; 24 in the root canal and 28 in the tooth, but only 8 were common to both samples. By culture, 23 taxa were identified and 27 by PCR cloning, but only 5 were common to both methods. The culture method revealed 16 bacterial species in the root canal compared with 11 in the tooth, but only 4 were common to both samples. This simple experiment illustrates the potential for sampling and detection methods to introduce distortions in identification of the root canal flora.
Table 2. Advantages and disadvantages of molecular-based methods
| Advantages | Disadvantages |
| Assesses DNA sequences (target-specific with PCR) | Risk of contamination |
| Potential to recover majority of microbes | Relies on established sequence identities |
| Modest skill level required | Relies on well-designed primer set |
| Relatively quick and straightforward processing | DNA recovery influenced by cell lysis method |
| Can freeze samples for later processing | Unable to distinguish live from dead cells |
| High specificity |
Why is there a disparity between the methods?
Historically, one of the most important challenges in determining microbial diversity has been that microorganisms are extracted from their environment and then studied microscopically, or in culture. There is clear evidence that not all organisms present in the root canals are subsequently cultivated, as revealed by microscopy of direct smears (27, 32, 85, 116) and by correlative light and electron microscopy (45).
The diversity of microbial communities is usually based on complex environmental, nutritional, and communicative interactions between species. By necessity, the process of taking a sample underestimates both the number and variety of species present in the original ecological niche (117, 118). The sampling method, conditions of transport, storage, culture (dilution, handling, media, incubation and atmosphere), and laboratory sub-culture all have the potential to bias the resulting types and numbers of species (119). At sampling, bacteria may be dormant, fragile, or in a starvation state, which may also influence recovery (120, 121).
Recovery and analysis of nucleotide sequences in a biological sample is completely independent of microbial viability or growth. By recovering available DNA, the molecular approach has an advantage over conventional culture because there is the potential to detect all culturable, previously uncultured, and not-yet-culturable micro-organisms in mixed samples (122). These methods also have drawbacks as each physical, chemical, and biological step may bias sequence recovery (123, 124). Whether the sample sequences are representative depends on the process of sample collection, transport, and efficacy of the DNA extraction. Successful isolation of DNA is influenced by physico-chemical factors in the sampled environment (e.g. inhibitors and nucleases) and biological properties of individual species (e.g. Gram-negative species are more susceptible than Gram-positive species to cell lysis) (123, 124). Some factors that influence the molecular analysis of the microbial flora in mixed samples are summarized in Table 3.
It should be pointed out that there are few studies which have evaluated samples from the same case by both culture and molecular approaches (67, 68, 125). In all such studies there is a disparity between the culture and molecular findings, where some species are identified by both approaches and some species are detected by one but not the other method. More work is needed in this area to satisfactorily reconcile, clarify, and resolve the reasons for different results from these two approaches.
Micro-organisms involved in post-treatment disease
The goal of instrumentation and irrigation of the root canal system is infection control. However, limitations in access to micro-organisms and in treatment efficacy mean that conventional root canal treatment falls short of this goal and low numbers of bacteria may survive in 30–50% of cases (7, 13, 14, 134–138). Application of an inter-appointment antibacterial dressing in multi-visit treatment does improve bactericidal efficacy (134, 137, 139–141) before canal filling. The root filling can entomb residual bacteria and deprive them of nutrients, and block direct communication with the periapical tissues. Persistent periapical disease requires that residual bacteria maintain communication with the periapical tissues and are capable of eliciting inflammation. Therefore, a distinction is made between species that survive root canal treatment and species involved in post-treatment disease (Fig. 1).
Table 3. Some factors that influence molecular analysis of micro-organisms in mixed samples
| Co-extraction of PCR inhibitors (humic acids, polysaccharides) with nucleic acids (124) |
| Changes in composition of 16S rDNA clone libraries caused by environmental DNA concentration (126) |
| G+C composition of 16S rRNA genes (127) |
| Genome size and copy number of rRNA genes (128, 129) |
| Lack of information on genome size and gene copy number for uncultured micro-organisms (124) |
| Choice of PCR primers, their design, and number of replication cycles (130) |
| Differential cloning efficiencies of PCR amplicons (123) |
| Formation of mosaic sequences from separate genes (131) |
| Limitations in quality and integrity of sequence databases (132, 133) |
Many studies have described bacteria present in post-treatment samples and there is a clear tendency for Gram-positive species to prevail after antimicrobial endodontic treatment (7, 9, 11, 12, 142). While there are no particular definitive resistant micro-organisms, certain species are reported in greater frequency as survivors after instrumentation, in particular but not exclusively, Streptococcus, Propionibacterium, Eubacterium and Actinomyces species (9, 143).
Will all species live and maintain a periapical inflammation?
Some bacteria may survive chemomechanical treatment but do not have the capacity to induce or maintain apical periodontitis. Based on culture studies of samples taken after instrumentation and irrigation, there are no recoverable bacteria in 50–70% of canals (7, 134–136, 138, 143). The converse of this is that 30–50% of cases will have recoverable bacteria at the time of root filling, yet not all of these cases end in post-treatment disease. In one study, recoverable bacteria were present in 40% of teeth at the time of root filling, yet in those cases with infection, 68% healed (9). This demonstrates that the presence of infection at the time of root filling does not always result in post-treatment disease. This was confirmed histologically in a study evaluating the impact of contemporary root canal treatment procedures on the microbiota in the mesial roots of mandibular molars with periapical lesions; it was found that 14/16 roots still demonstrated residual biofilms in the apical complexities after single-visit treatment (16).
Since the success rate of single-visit root canal treatment is in the range of 70–85%, it raises the question of the fate of the residual microbiota in cases that heal. Entombed in the filled canal, they may die, but in some cases residual bacteria may continue to interact with the host tissues, resulting in a slow healing response over many years (144–146). In other cases, it is reasonable to think that some species endure the low nutrient or starvation conditions and survive in the filled root canal, which is consistent with a less favorable healing outcome in cases where infection is present at the time of canal filling (9, 147). Without access to the periapical tissue, the lesion would likely heal; however, there remains a risk of persistent apical periodontitis if microbes survive and have an opportunity to communicate with the periapical tissues.
Intraradicular infection
In untreated cases, analysis of the root canal infection depends on careful, contamination-free access to sample the microbiota, which typically contain high cell numbers of multiple species. In studies of infected root-filled teeth, a significant challenge is retrieval of the microbial flora, usually in low numbers, from the filled root canal system. Removal of the root filling by mechanical instrumentation may generate heat and the use of solvents (usually chloroform) may kill remaining bacteria. One study compared the impact of using chloroform on the recovery of bacteria and found that 78% of the canals were positive for bacteria without chloroform while only 48% were positive with its use (148). Some studies specifically exclude the use of chloroform before microbiological examination for this reason (149–153).
In general, the bacterial species recovered from root-filled teeth are mostly a subset of those found in untreated teeth, but with a reduced species diversity and quantity. Typically, Gram-positive facultative bacteria dominate the microbiota (81, 148, 153). In poorly treated teeth, the microbial flora resemble those recovered from untreated teeth (149, 153–156).
A consistent finding across multiple studies is the frequent isolation of E. faecalis (81, 148, 150–154, 156–160), although some studies failed to identify it (67, 149) and some studies describe the presence of E. faecalis in a small proportion of cases (161, 162). The presence of E. faecalis is notable not only because of a high prevalence in previously root-filled teeth, but it is usually infrequently found in teeth with primary infection (7, 72, 163). This apparent paradox has raised questions about how E. faecalis has come to be present in a high proportion of teeth after, but not before, root filling (based on cross-sectional studies) (21, 164). One likely contributory factor is the operative events during the period of the original treatment. E. faecalis has been reported in a significantly higher proportion of cases when there has been protracted treatment or the canals have been left open (165). Another question is whether the source is resident in the host oral microflora or is of transient derivation, and a recent study points to an exogenous source of E. faecalis (166). When present in teeth with post-treatment disease, it is more difficult to eradicate during subsequent re-treatment (153).
Other species that appear with greater prevalence in root-filled teeth with post-treatment disease include Streptococcus species (148–150, 152–154), Candida species (predominantly C. albicans) (148–150, 152–154, 156, 167, 168), and Propionibacterium species (169, 170). In culture-independent studies, there are reports of Filifactor alocis and Dialister species (156, 160, 170, 171).
In most studies, there is a reduced diversity of isolated species when compared to the microbiota of untreated teeth. In culture-based studies, there is a higher incidence of monoinfections, particularly of E. faecalis (148, 150, 151, 153), although molecular-based studies report a greater number of species (170, 172).
Extraradicular infection
The periapical granuloma is in intimate anatomical contact with the root apex and frequently communicates with the apical root canal, so obtaining a bacteriological sample from the periapical tissues without contamination from either the root apex or canal presents an intricate challenge. It is also the source of controversy in this area. Almost 50 years ago Möller demonstrated that, with a conventional surgical approach using routine isolation and suction, it was impossible to recover periapical tissue without contamination (81). Contamination control samples from adjacent soft tissue and bone were repeatedly positive, which invalidated the targeted periapical tissue sample. He solved the problem by stringent isolation using a custom-made acrylic shield, which allowed him to work and take samples without contamination from the surrounding field (81). It is important to recognize the significance of this accomplishment because bacteriological studies using root-end scrapings or samples including the root tip, surgical samples without stringent isolation, and periapical tissue sampling via the root canal are subject to an unacceptable risk of contamination. If the overlying soft tissue is carefully isolated, cleaned, and disinfected, it is possible to successfully take samples by needle aspiration of the periapical granuloma.
The interaction of root canal microbes with the host occurs in a dynamic interplay at the root apex. Small groups of bacteria may breach the apical foramen and the host’s normal immune response is to detect and destroy invading bacteria. This is the role and function of the periapical granuloma. In some cases, bacteria may interfere with the host defense and overcome it, which is the reason for the presention of acute symptoms such as pain, swelling, or exudation. The vast majority of bacteria succumb to the host response, occasionally with adjunctive clinical support in acute cases, and are therefore not generally considered a sustainable form of extraradicular infection.
Only a few oral species have a demonstrated capacity for establishment and long-term survival in host tissue, independent of the root canal system. Verification that the infection is extraradicular and unconnected with the root canal system in such cases is borne out by correlative light and electron microscopy of serial sections, illustrated by a tooth with a periapical actinomycosis (173). This need not exclude some cases where there may be a contiguous arrangement between Actinomyces species in the granuloma and apical root canal. The clinical significance of the extraradicular location is that conventional root canal treatment will not address the source of the persisting infection. Periapical surgery is required to remove it.
The main genera involved in extraradicular infections are Actinomyces species and Propionibacterium species (174–176). The presence of A. israelii is a recurrent finding in therapy-resistant cases (72, 163, 176) and is the most common species involved in actinomycosis (177). Of the Actinomyces species, A. israelii is isolated at twice the prevalence of Actinomyces gerencseriae (formerly A. israelii serotype II) in human abscesses (177) but in half the prevalence in primary root canal infections (163). The role of A. gerencseriae in persistent infection after root filling remains to be clarified. Another Actinomyces species, Actinomyces radicidentis, is occasionally associated with post-treatment disease (169, 178).
Environmental selection
Lourens Baas Becking, a Dutch microbiologist, is credited with the statement “Everything is everywhere, but the environment selects,” which refers to the local environment as a critical determinant in microbial ecology and survival. In the oral cavity, distinct microenvironments at various soft and hard tissue surfaces influence the composition of the microbiota. The untreated infected root canal system is an environment that provides micro-organisms with a rich source of nutrition. Initially, there may be a source of carbohydrates facilitating growth of facultative anaerobes, but as the infection develops over time, the nutrients are mainly peptides and amino acids, which favor anaerobic proteolytic species.
In the filled root canal, most of the nutrients have been stripped away during earlier chemomechanical cleaning, leaving comparatively barren conditions for surviving microbial cells. These microbes generally face a low-nutrient or starvation environment, but in favorable conditions may encounter a serum-like fluid transudate from the periapical tissue.
Some species and individual strains are better equipped to endure periods of starvation, which enhances their long-term survival capacity. Species that are more prevalent in post-treatment disease, such as E. faecalis and C. albicans, have been shown to have a superior starvation survival capacity (18, 19) compared with strict anaerobes that dominate the microbiota in untreated cases (20), which helps explain why the latter group rarely participates in post-treatment disease.
In the extraradicular environment, cellular and humoral factors are deployed by the host to routinely eliminate micro-organisms that invade the periapical tissue. An ability of the micro-organism to strategically respond to the host defense is crucial for survival. The microbial arsenal includes evasion by physical concealment or biological evasion of host surveillance, cell-mediated phagocytosis, or evasion of immune protein-mediated antibodies and complement. Species involved with extraradicular infection, such as A. israelii and A. radicidentis, have been shown to survive in host tissue in animal experiments (179–183) through bacterial cohesion and avoidance of phagocytosis.
There are many further factors that constitute the microenvironment such as the host surface and local defense, microanatomy, anaerobiosis, pH, etc. Much more work is needed in this area and discovery of specific environmental factors that regulate local microbial ecology should open up therapeutic options which are more precisely directed at particular types of biofilms and sites.
Conclusion
The effectiveness of root canal treatment depends on microbial control through a combination of direct bactericidal and ecologic effects. Hardier species that have a greater capacity to resist antimicrobial treatment and that adapt to a nutrient-depleted environment are best suited to endure in the root-filled canal. Where microbes survive, facultative Gram-positive bacteria dominate the remaining bacterial communities.
Post-treatment disease is primarily due to persisting (or possibly recurrent) infection of the apical part of the root canal system. An ineffective first attempt at root canal treatment may leave a microbiota that is more difficult to eradicate and located in anatomically complex regions of the root canal system.
The re-treatment strategy for post-treatment disease may depend on the case type. In teeth with inadequate previous treatment, such as missed canals, the infection resembles an untreated case and it should respond well to conventional root canal treatment. In cases of sub-standard treatment, the infection is likely dominated by Gram-positive species, which may be more resistant to re-treatment. In well-treated teeth, post-treatment disease may be due to persistent intra- or extraradicular infection and, in addition to a conventional chemomechanical approach, consideration should be given to surgical intervention. The treatment priority remains the same in all cases: to obtain access to the apical root canal anatomy and control residual infection.
Healing outcome data indicates that around 85% of periapical lesions resolve after root canal treatment of teeth with primary infections. In cases requiring re-treatment, about 75% of periapical lesions resolve over time. Taken collectively, conventional root canal treatment offers an excellent outcome in terms of the periapical healing rate. Nevertheless, many important questions remain. Why some microbes are able to participate in post-treatment disease in some cases is still not well understood. The contribution of microbial dynamics and pathogenicity, specific anatomical attributes, and the role of the host defense are important factors that await clarification.
Over the past two decades, there have been many advances in instrumentation techniques, materials, and technology. The widespread adoption of the operating microscope has meant that the clinician can see the task with far greater visual acuity. Future improvements in root canal treatment will likely come from a deeper biological insight into the microbial pathogenicity and the factors regulating community behavior.
References
1. Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol 2004: 12: 129–134.
2. Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology 2003: 149: 279–294.
3. Eriksen HM. Epidemiology of apical periodontitis. In: Ørstavik D, Pitt Ford TR, eds. Essential Endodontology. Prevention and Treatment of Apical Periodontitis. Blackwell Science, 1998: 179–191.
4. ADA: 2005–06 Survey of Dental Services Rendered. American Dental Association, 2007: 1–167.
5. Figdor D. Apical periodontitis: a very prevalent problem. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2002: 94: 651–652.
6. Möller ÅJR, Fabricius L, Dahlén G, Sundqvist G, Happonen RP. Apical periodontitis development and bacterial response to endodontic treatment. Experimental root canal infections in monkeys with selected bacterial strains. Eur J Oral Sci 2004: 112: 207–215.
7. Byström A, Sundqvist G. The antibacterial action of sodium hypochlorite and EDTA in 60 cases of endodontic therapy. Int Endod J 1985: 18: 35–40.
8. Gomes BP, Lilley JD, Drucker DB. Variations in the susceptibilities of components of the endodontic microflora to biomechanical procedures. Int Endod J 1996: 29: 235–241.
9. Sjögren U, Figdor D, Persson S, Sundqvist G. Influence of infection at the time of root filling on the outcome of endodontic treatment of teeth with apical periodontitis. Int Endod J 1997: 30: 297–306 [Published erratum appears in Int Endod J 1998: 31: 148].
10. Chu FC, Leung WK, Tsang PC, Chow TW, Samaranayake LP. Identification of cultivable microorganisms from root canals with apical periodontitis following two-visit endodontic treatment with antibiotics/steroid or calcium hydroxide dressings. J Endod 2006: 32: 17–23.
11. Sakamoto M, Siqueira JF Jr, Rôças IN, Benno Y. Bacterial reduction and persistence after endodontic treatment procedures. Oral Microbiol Immunol 2007: 22: 19–23.
12. Siqueira JF Jr, Paiva SS, Rôças IN. Reduction in the cultivable bacterial populations in infected root canals by a chlorhexidine-based antimicrobial protocol. J Endod 2007: 33: 541–547.
13. Byström A, Sundqvist G. Bacteriologic evaluation of the efficacy of mechanical root canal instrumentation in endodontic therapy. Scand J Dent Res 1981: 89: 321–328.
14. Byström A, Sundqvist G. Bacteriologic evaluation of the effect of 0.5 percent sodium hypochlorite in endodontic therapy. Oral Surg Oral Med Oral Pathol 1983: 55: 307–312.
15. Matsuo T, Shirakami T, Ozaki K, Nakanishi T, Yumoto H, Ebisu S. An immunohistological study of the localization of bacteria invading root pulpal walls of teeth with periapical lesions. J Endod 2003: 29: 194–200.
16. Nair PNR, Henry S, Cano V, Vera J. Microbial status of apical root canal system of human mandibular first molars with primary apical periodontitis after “one-visit” endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2005: 99: 231–252.
17. Figdor D, Sundqvist G. A big role for the very small—understanding the endodontic microbial flora. Aust Dent J 2007: 52: S38–S51.
18. Figdor D, Davies JK, Sundqvist G. Starvation survival, growth and recovery of Enterococcus faecalis in human serum. Oral Microbiol Immunol 2003: 18: 234–239.
19. Richards D, Davies JK, Figdor D. Starvation survival and recovery in serum of Candida albicans compared with Enterococcus faecalis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2010: 110: 125–130.
20. Brundin M, Figdor D, Sundqvist G, Sjögren U. Starvation response and growth in serum of Fusobacterium nucleatum, Peptostreptococcus anaerobius, Prevotella intermedia, and Pseudoramibacter alactolyticus. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2009: 108: 129–134.
21. Sundqvist G, Figdor D. Life as an endodontic pathogen. Ecological differences between the untreated and root-filled root canals. Endod Topics 2003: 6: 3–28.
22. Ray HA, Trope M. Periapical status of endodontically treated teeth in relation to the technical quality of the root filling and the coronal restoration. Int Endod J 1995: 28: 12–18.
23. Hommez GM, Coppens CR, De Moor RJ. Periapical health related to the quality of coronal restorations and root fillings. Int Endod J 2002: 35: 680–689.
24. Tavares PB, Bonte E, Boukpessi T, Siqueira JF Jr, Lasfargues JJ. Prevalence of apical periodontitis in root canal-treated teeth from an urban French population: influence of the quality of root canal fillings and coronal restorations. J Endod 2009: 35: 810–813.
25. Ricucci D, Bergenholtz G. Bacterial status in root-filled teeth exposed to the oral environment by loss of restoration and fracture or caries—a histobacteriological study of treated cases. Int Endod J 2003: 36: 787–802.
26. Koch R. Die Aetiologie der Milzbrand-Krakheit, begründet auf die Entwicklungsgeschichte des Bacillus Anthracis. Beiträge zur Biologie der Pflanzen 1876: 2: 277–310.
27. Miller WD. An introduction to the study of the bacterio-pathology of the dental pulp. The Dental Cosmos 1894: 36: 505–527.
28. Siqueira JF Jr, Rôças IN. Diversity of endodontic microbiota revisited. J Dent Res 2009: 88: 969–981.
29. Pupo GM, Karaolis DK, Lan R, Reeves PR. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun 1997: 65: 2685–2692.
30. Takahashi K. Microbiological, pathological, inflammatory, immunological and molecular biological aspects of periradicular disease. Int Endod J 1998: 31: 311–325.
31. Bowden GH. Oral biofilms: an archive of past events? In: Newman HN, Wilson M, eds. Dental Plaque Revisited—Oral Biofilms in Health and Disease. Bioline, 1999: 211–235.
32. Crawford JJ, Shankle RJ. Application of newer methods to study the importance of root canal and oral microbiota in endodontics. Oral Surg Oral Med Oral Pathol 1961: 14: 1109–1123.
33. Sundqvist G. Taxonomy, ecology, and pathogenicity of the root canal flora. Oral Surg Oral Med Oral Pathol 1994: 78: 522–530.
34. Fabricius L, Dahlén G, Holm SE, Möller ÅJR. Influence of combinations of oral bacteria on periapical tissues of monkeys. Scand J Dent Res 1982: 90: 200–206.
35. Fabricius L, Dahlén G, Öhman AE, Möller ÅJR. Predominant indigenous oral bacteria isolated from infected root canals after varied times of closure. Scand J Dent Res 1982: 90: 134–144.
36. Möller ÅJR, Fabricius L, Dahlén G, Öhman AE, Heyden G. Influence on periapical tissues of indigenous oral bacteria and necrotic pulp tissue in monkeys. Scand J Dent Res 1981: 89: 475–484.
37. Allard U, Strömberg U, Strömberg T. Endodontic treatment of experimentally induced apical periodontitis in dogs. Endod Dent Traumatol 1987: 3: 240–244.
38. Tani-Ishii N, Wang CY, Tanner A, Stashenko P. Changes in root canal microbiota during the development of rat periapical lesions. Oral Microbiol Immunol 1994: 9: 129–135.
39. Sundqvist GK, Eckerbom MI, Larsson ÅP, Sjögren UT. Capacity of anaerobic bacteria from necrotic dental pulps to induce purulent infections. Infect Immun 1979: 25: 685–693.
40. Ribeiro Sobrinho AP, de Melo Maltos SM, Farias LM, de Carvalho MA, Nicoli JR, de Uzeda M, Vieira LQ. Cytokine production in response to endodontic infection in germ-free mice. Oral Microbiol Immunol 2002: 17: 344–353.
41. Ehrlich GD, Ahmed A, Earl J, Hiller NL, Costerton JW, Stoodley P, Post JC, DeMeo P, Hu FZ. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol Med Microbiol 2010: 59: 269–279.
42. Ehrlich GD, Hu FZ, Shen K, Stoodley P, Post JC. Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin Orthop Relat Res 2005: 20–24.
43. Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol 2005: 13: 589–595.
44. Kohsaka T, Kumazawa M, Yamasaki M, Nakamura H. Periapical lesions in rats with streptozotocin-induced diabetes. J Endod 1996: 22: 418–421.
45. Nair PNR. Light and electron microscopic studies of root canal flora and periapical lesions. J Endod 1987: 13: 29–39.
46. Richardson N, Mordan NJ, Figueiredo JA, Ng YL, Gulabivala K. Microflora in teeth associated with apical periodontitis: a methodological observational study comparing two protocols and three microscopy techniques. Int Endod J 2009: 42: 908–921.
47. Kolter R. Surfacing views of biofilm biology. Trends Microbiol 2005: 13: 1–2.
48. Flynn D, Ready DR, Mordan N, Pratten J, Gulabivala K. The effect of conditioning films on colonisation of root canal dentine by endodontic bacterial isolates. Int Endod J 2008: 41: 815.
49. Kolenbrander PE, Egland PG, Diaz PI, Palmer RJ Jr. Genome–genome interactions: bacterial communities in initial dental plaque. Trends Microbiol 2005: 13: 11–15.
50. de Beer D, Stoodley P, Lewandowski Z. Liquid flow in heterogeneous biofilms. Biotechnol Bioeng 1994: 44: 636–641.
51. Juhas M, Eberl L, Tummler B. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol 2005: 7: 459–471.
52. Suntharalingam P, Cvitkovitch DG. Quorum sensing in streptococcal biofilm formation. Trends Microbiol 2005: 13: 3–6.
53. Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 2003: 112: 1466–1477.
54. Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol 2005: 13: 20–26.
55. Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 2004: 230: 13–18.
56. Lewis K. Persister cells. Annu Rev Microbiol 2010: 64: 357–372.
57. Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol 2005: 13: 7–10.
58. Rojekar S, Mordan N, Ng Y-L, Figueiredo JAP, Gulabivala K. In situ immunocytochemical colloidal gold probing of three bacterial species in the root canal system of teeth associated with apical periodontitis. Int Endod J 2006: 39: 739.
59. Iacovidou A, Mordan N, Ready D, Figueiredo JAP, Gulabivala K. Utility of the FISH technique in conjunction with confocal microscopy to study the intraradicular microflora of teeth associated with apical periodontitis. Int Endod J 2008: 41: 818.
60. Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol 2005: 13: 34–40.
61. Moore WEC, Moore LVH. The bacteria of periodontal diseases. Periodontol 2000 1994: 5: 66–77.
62. Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol 2001: 183: 3770–3783.
63. Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000 2006: 42: 80–87.
64. Kantz WE, Henry CA. Isolation and classification of anaerobic bacteria from intact pulp chambers of non-vital teeth in man. Archs Oral Biol 1974: 19: 91–96.
65. Wittgow WC Jr, Sabiston CB Jr. Microorganisms from pulpal chambers of intact teeth with necrotic pulps. J Endod 1975: 1: 168–171.
66. Sundqvist G. Bacteriological studies of necrotic dental pulps. Umeå University Odontological Dissertations No. 7. Department of Oral Microbiology, Umeå University, Sweden, 1976.
67. Rolph HJ, Lennon A, Riggio MP, Saunders WP, MacKenzie D, Coldero L, Bagg J. Molecular identification of microorganisms from endodontic infections. J Clin Microbiol 2001: 39: 3282–3289.
68. Munson MA, Pitt-Ford T, Chong B, Weightman A, Wade WG. Molecular and cultural analysis of the microflora associated with endodontic infections. J Dent Res 2002: 81: 761–766.
69. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 2005: 43: 5721–5732.
70. Gomes BP, Drucker DB, Lilley JD. Positive and negative associations between bacterial species in dental root canals. Microbios 1994: 80: 231–243.
71. Rôças IN, Siqueira JF Jr. Root canal microbiota of teeth with chronic apical periodontitis. J Clin Microbiol 2008: 46: 3599–3606.
72. Sundqvist G. Associations between microbial species in dental root canal infections. Oral Microbiol Immunol 1992: 7: 257–262.
73. Grenier D, Mayrand D. Nutritional relationships between oral bacteria. Infect Immun 1986: 53: 616–620.
74. Marsh PD. Host defenses and microbial homeostasis: role of microbial interactions. J Dent Res 1989: 68: 1567–1575.
75. Sundqvist G. Ecology of the root canal flora. J Endod 1992: 18: 427–430.
76. Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Ann Rev Microbiol 2002: 56: 117–137.
77. Loesche WJ. Importance of nutrition in gingival crevice microbial ecology. Periodontics 1968: 6: 245–249.
78. Love RM. Enterococcus faecalis—a mechanism for its role in endodontic failure. Int Endod J 2001: 34: 399–405.
79. ter Steeg PF, van der Hoeven JS, de Jong MH, van Munster PJ, Jansen MJ. Enrichment of subgingival microflora on human serum leading to accumulation of Bacteroides species, Peptostreptococci and Fusobacteria. Antonie van Leeuwenhoek 1987: 53: 261–272.
80. ter Steeg PF, van der Hoeven JS. Development of periodontal microflora on human serum. Microb Ecol Health Dis 1989: 2: 1–10.
81. Möller ÅJR. Microbiological examination of root canals and periapical tissues of human teeth. Methodological studies. Odontol Tidsk 1966: 74(Suppl): 1–380.
82. Keudell K, Conte M, Fujimoto L, Ernest M, Berry HG. Microorganisms isolated from pulp chambers. J Endod 1976: 2: 146–148.
83. Holt JG. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins, 1994.
84. Brown LR, Rudolph CE. Isolation and identification of microorganisms from unexposed canals of pulp-involved teeth. Oral Surg 1957: 10: 1094–1099.
85. Engström B, Frostell G. Bacteriological studies of the non-vital pulp in cases with intact pulp cavities. Acta Odont Scand 1961: 19: 23–39.
86. Holdeman LV, Cato EP, Moore WEC. Anaerobe Laboratory Manual. VPI Anaerobe Laboratory, Virginia Polytechnic Institute and State University, 1977.
87. Goodman AD. Isolation of anaerobic bacteria from the root canal systems of necrotic teeth by the use of a transport solution. Oral Surg Oral Med Oral Pathol 1977: 43: 766–770.
88. Kipioti A, Nakou M, Legakis N, Mitsis F. Microbiological findings of infected root canals and adjacent periodontal pockets in teeth with advanced periodontitis. Oral Surg Oral Med Oral Pathol 1984: 58: 213–220.
89. Gomes BPFA, Drucker DB, Lilley JD. Association of specific bacteria with some endodontic signs and symptoms. Int Endod J 1994: 27: 291–298.
90. Hoshino E, Ando N, Sato M, Kota K. Bacterial invasion of non-exposed dental pulp. Int Endod J 1992: 25: 2–5.
91. Wasfy MO, McMahon KT, Minah GE, Falkler WA Jr. Microbiological evaluation of periapical infections in Egypt. Oral Microbiol Immunol 1992: 7: 100–105.
92. Weiger R, Manncke B, Werner H, Löst C. Microbial flora of sinus tracts and root canals of non-vital teeth. Endod Dent Traumatol 1995: 11: 15–19.
93. Hirai K, Tagami A, Okuda K. Isolation and classification of anaerobic bacteria from pulp cavities of nonvital teeth in man. Bull Tokyo Dent Coll 1991: 32: 95–98.
94. Hashioka K, Yamasaki M, Nakane A, Horiba N, Nakamura H. The relationship between clinical symptoms and anaerobic bacteria from infected root canals. J Endod 1992: 18: 558–561.
95. Brauner AW, Conrads G. Studies into the microbial spectrum of apical periodontitis. Int Endod J 1995: 28: 244–248.
96. Le Goff A, Bunetel L, Mouton C, Bonnaure-Mallet M. Evaluation of root canal bacteria and their antimicrobial susceptibility in teeth with necrotic pulp. Oral Microbiol Immunol 1997: 12: 318–322.
97. Lana MA, Ribeiro-Sobrinho AP, Stehling R, Garcia GD, Silva BK, Hamdan JS, Nicoli JR, Carvalho MA, Farias Lde M. Microorganisms isolated from root canals presenting necrotic pulp and their drug susceptibility in vitro. Oral Microbiol Immunol 2001: 16: 100–105.
98. Gulabivala K, Spratt DA, McNab R, Ng Y-L, Ready D, Wilson M. Species-richness of Gram-positive coccoid morphotypes from root canals determined by phenotypic and genetic measures. Int Endod J 2005: 38: 918.
99. Conrads G, Gharbia SE, Gulabivala K, Lampert F, Shah HN. The use of a 16S rDNA directed PCR for the detection of endodontopathogenic bacteria. J Endod 1997: 23: 433–438.
100. Jung IY, Choi BK, Kum KY, Roh BD, Lee SJ, Lee CY, Park DS. Molecular epidemiology and association of putative pathogens in root canal infection. J Endod 2000: 26: 599–604.
101. Fouad AF, Barry J, Caimano M, Clawson M, Zhu Q, Carver R, Hazlett K, Radolf JD. PCR-based identification of bacteria associated with endodontic infections. J Clin Microbiol 2002: 40: 3223–3231.
102. Siqueira JF Jr, Rôças IN, Moraes SR, Santos KR. Direct amplification of rRNA gene sequences for identification of selected oral pathogens in root canal infections. Int Endod J 2002: 35: 345–351.
103. Gonçalves RB, Mouton C. Molecular detection of Bacteroides forsythus in infected root canals. J Endod 1999: 25: 336–340.
104. Siqueira Jr JF, Rôças IN, Souto R, de Uzeda M, Colombo AP. Checkerboard DNA–DNA hybridization analysis of endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2000: 89: 744–748.
105. Rôças IN, Siqueira JF Jr, Santos KR, Coelho AM. “Red complex” (Bacteroides forsythus, Porphyromonas gingivalis, and Treponema denticola) in endodontic infections: a molecular approach. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001: 91: 468–471.
106. Baumgartner JC, Watkins BJ, Bae KS, Xia T. Association of black-pigmented bacteria with endodontic infections. J Endod 1999: 25: 413–415.
107. Siqueira JF Jr, Róças IN, Oliveira JC, Santos KR. Molecular detection of black-pigmented bacteria in infections of endodontic origin. J Endod 2001: 27: 563–566.
108. Siqueira JF Jr, Rôças IN, Favieri A, Santos KR. Detection of Treponema denticola in endodontic infections by 16S rRNA gene-directed polymerase chain reaction. Oral Microbiol Immunol 2000: 15: 335–337.
109. Jung IY, Choi B, Kum KY, Yoo YJ, Yoon TC, Lee SJ, Lee CY. Identification of oral spirochetes at the species level and their association with other bacteria in endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001: 92: 329–334.
110. Siqueira JF Jr, Rôças IN, Favieri A, Oliveira JC, Santos KR. Polymerase chain reaction detection of Treponema denticola in endodontic infections within root canals. Int Endod J 2001: 34: 280–284.
111. Hashimura T, Sato M, Hoshino E. Detection of Slackia exigua, Mogibacterium timidum and Eubacterium saphenum from pulpal and periradicular samples using the Polymerase Chain Reaction (PCR) method. Int Endod J 2001: 34: 463–470.
112. Molander A, Lundquist P, Papapanou PN, Dahlén G, Reit C. A protocol for polymerase chain reaction detection of Enterococcus faecalis and Enterococcus faecium from the root canal. Int Endod J 2002: 35: 1–6.
113. Siqueira Jr JF, Rôças IN, De Uzeda M, Colombo AP, Santos KR. Comparison of 16S rDNA-based PCR and checkerboard DNA–DNA hybridisation for detection of selected endodontic pathogens. J Med Microbiol 2002: 51: 1090–1096.
114. Siqueira JF Jr, Rôças IN, Paiva SS, Magalhães KM, Guimarães-Pinto T. Cultivable bacteria in infected root canals as identified by 16S rRNA gene sequencing. Oral Microbiol Immunol 2007: 22: 266–271.
115. Kumar T, Spratt DA, Ng Y-L, Gulabivala K. A preliminary evaluation of a new method for sampling the intra-radicular bacterial flora. Int Endod J 2002: 35: 85.
116. Sulitzeanu A, Beutner EH, Epstein LI. Bacteriologic studies of pulp-involved teeth by cultural and microscopic methods. J Am Dent Assoc 1964: 69: 300–307.
117. Brock TD. The study of microorganisms in situ: progress and problems. 41st Symposium, Society for General Microbiology, Ecology of Microbial Communities; Cambridge University Press, 1987: 1–17.
118. Schlegel HG, Jannasch HW. Prokaryotes and their habitats. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH, eds. The Prokaryotes. Springer Verlag, 1991: 1: 75–125.
119. O’Donnell AG, Goodfellow M, Hawksworth DL. Theoretical and practical aspects of the quantification of biodiversity among microorganisms. Philos Trans R Soc Lond B Biol Sci 1994: 345: 65–73.
120. Rollins DM, Colwell RR. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl Environ Microbiol 1986: 52: 531–538.
121. Roszak DB, Grimes DJ, Colwell RR. Viable but nonrecoverable stage of Salmonella enteritidis in aquatic systems. Can J Microbiol 1984: 30: 334–338.
122. Pace NR. A molecular view of microbial diversity and the biosphere. Science 1997: 276: 734–740.
123. Embley TM, Stackebrandt E. The molecular phylogeny and systematics of the Actinomycetes. Annu Rev Microbiol 1994: 48: 257–289.
124. von Wintzingerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 1997: 21: 213–229.
125. Pratten J, Wilson M, Spratt DA. Characterization of in vitro oral bacterial biofilms by traditional and molecular methods. Oral Microbiol Immunol 2003: 18: 45–49.
126. Chandler DP, Fredrickson JK, Brockman FJ. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol 1997: 6: 475–482.
127. Reysenbach AL, Giver LJ, Wickham GS, Pace NR. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol 1992: 58: 3417–3418.
128. Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev 1990: 54: 502–539.
129. Farrelly V, Rainey FA, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol 1995: 61: 2798–2801.
130. Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 1996: 62: 625–630.
131. Borriello F, Krauter KS. Reactive site polymorphism in the murine protease inhibitor gene family is delineated using a modification of the PCR reaction (PCR+1). Nucleic Acids Res 1990: 18: 5481–5487.
132. Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol 1992: 174: 5072–5078.
133. Stackebrandt E, Liesack W, Goebel BM. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J 1993: 7: 232–236.
134. Shuping GB, Ørstavik D, Sigurdsson A, Trope M. Reduction of intracanal bacteria using nickel–titanium rotary instrumentation and various medications. J Endod 2000: 26: 751–755.
135. Kvist T, Molander A, Dahlén G, Reit C. Microbiological evaluation of one- and two-visit endodontic treatment of teeth with apical periodontitis: a randomized, clinical trial. J Endod 2004: 30: 572–576.
136. Vianna ME, Horz HP, Gomes BP, Conrads G. In vivo evaluation of microbial reduction after chemo-mechanical preparation of human root canals containing necrotic pulp tissue. Int Endod J 2006: 39: 484–492.
137. Siqueira JF Jr, Guimarães-Pinto T, Rôças IN. Effects of chemomechanical preparation with 2.5% sodium hypochlorite and intracanal medication with calcium hydroxide on cultivable bacteria in infected root canals. J Endod 2007: 33: 800–805.
138. Siqueira JF Jr, Magalhães KM, Rôças IN. Bacterial reduction in infected root canals treated with 2.5% NaOCl as an irrigant and calcium hydroxide/camphorated paramonochlorophenol paste as an intracanal dressing. J Endod 2007: 33: 667–672.
139. Byström A, Claesson R, Sundqvist G. The antibacterial effect of camphorated paramonochlorophenol, camphorated phenol and calcium hydroxide in the treatment of infected root canals. Endod Dent Traumatol 1985: 1: 170–175.
140. Sjögren U, Figdor D, Spångberg L, Sundqvist G. The antimicrobial effect of calcium hydroxide as a short-term intracanal dressing. Int Endod J 1991: 24: 119–125.
141. McGurkin-Smith R, Trope M, Caplan D, Sigurdsson A. Reduction of intracanal bacteria using GT rotary instrumentation, 5.25% NaOCl, EDTA, and Ca(OH)2. J Endod 2005: 31: 359–363.
142. Chávez de Paz LE, Molander A, Dahlén G. Gram-positive rods prevailing in teeth with apical periodontitis undergoing root canal treatment. Int Endod J 2004: 37: 579–587.
143. Siqueira JF Jr, Rôças IN, Paiva SS, Guimarães-Pinto T, Magalhães KM, Lima KC. Bacteriologic investigation of the effects of sodium hypochlorite and chlorhexidine during the endodontic treatment of teeth with apical periodontitis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2007: 104: 122–130.
144. Molven O, Halse A, Fristad I, MacDonald-Jankowski D. Periapical changes following root-canal treatment observed 20–27 years postoperatively. Int Endod J 2002: 35: 784–790.
145. Ng YL, Mann V, Rahbaran S, Lewsey J, Gulabivala K. Outcome of primary root canal treatment: systematic review of the literature—Part 1. Effects of study characteristics on probability of success. Int Endod J 2007: 40: 921–939.
146. Ng YL, Mann V, Rahbaran S, Lewsey J, Gulabivala K. Outcome of primary root canal treatment: systematic review of the literature—Part 2. Influence of clinical factors. Int Endod J 2008: 41: 6–31.
147. Fabricius L, Dahlén G, Sundqvist G, Happonen RP, Möller ÅJR. Influence of residual bacteria on periapical tissue healing after chemomechanical treatment and root filling of experimentally infected monkey teeth. Eur J Oral Sci 2006: 114: 278–285.
148. Molander A, Reit C, Dahlén G, Kvist T. Microbiological status of root-filled teeth with apical periodontitis. Int Endod J 1998: 31: 1–7.
149. Cheung GS, Ho MW. Microbial flora of root canal-treated teeth associated with asymptomatic periapical radiolucent lesions. Oral Microbiol Immunol 2001: 16: 332–337.
150. Hancock HH 3rd, Sigurdsson A, Trope M, Moiseiwitsch J. Bacteria isolated after unsuccessful endodontic treatment in a North American population. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001: 91: 579–586.
151. Peciuliene V, Balciuniene I, Eriksen HM, Haapasalo M. Isolation of Enterococcus faecalis in previously root-filled canals in a Lithuanian population. J Endod 2000: 26: 593–595.
152. Peciuliene V, Reynaud AH, Balciuniene I, Haapasalo M. Isolation of yeasts and enteric bacteria in root-filled teeth with chronic apical periodontitis. Int Endod J 2001: 34: 429–434.
153. Sundqvist G, Figdor D, Persson S, Sjögren U. Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1998: 85: 86–93.
154. Pinheiro ET, Gomes BP, Ferraz CC, Sousa EL, Teixeira FB, Souza-Filho FJ. Microorganisms from canals of root-filled teeth with periapical lesions. Int Endod J 2003: 36: 1–11.
155. Rôças IN, Siqueira JF Jr, Aboim MC, Rosado AS. Denaturing gradient gel electrophoresis analysis of bacterial communities associated with failed endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2004: 98: 741–749.
156. Siqueira JF Jr, Rôças IN. Polymerase chain reaction-based analysis of microorganisms associated with failed endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2004: 97: 85–94.
157. Engström B. The significance of enterococci in root canal treatment. Odont Revy 1964: 15: 87–106.
158. Zoletti GO, Siqueira JF Jr, Santos KR. Identification of Enterococcus faecalis in root-filled teeth with or without periradicular lesions by culture-dependent and -independent approaches. J Endod 2006: 32: 722–726.
159. Sedgley C, Nagel A, Dahlén G, Reit C, Molander A. Real-time quantitative polymerase chain reaction and culture analyses of Enterococcus faecalis in root canals. J Endod 2006: 32: 173–177.
160. Gomes BP, Pinheiro ET, Jacinto RC, Zaia AA, Ferraz CC, Souza-Filho FJ. Microbial analysis of canals of root-filled teeth with periapical lesions using polymerase chain reaction. J Endod 2008: 34: 537–540.
161. Rôças IN, Hülsmann M, Siqueira JF Jr. Microorganisms in root canal-treated teeth from a German population. J Endod 2008: 34: 926–931.
162. Sakamoto M, Siqueira JF Jr, Rôças IN, Benno Y. Molecular analysis of the root canal microbiota associated with endodontic treatment failures. Oral Microbiol Immunol 2008: 23: 275–281.
163. Siqueira Jr JF, Rôças IN, Souto R, de Uzeda M, Colombo AP. Actinomyces species, streptococci, and Enterococcus faecalis in primary root canal infections. J Endod 2002: 28: 168–172.
164. Zehnder M, Guggenheim B. The mysterious appearance of enterococci in filled root canals. Int Endod J 2009: 42: 277–287.
165. Sirén EK, Haapasalo MP, Ranta K, Salmi P, Kerosuo EN. Microbiological findings and clinical treatment procedures in endodontic cases selected for microbiological investigation. Int Endod J 1997: 30: 91–95.
166. Vidana R, Sullivan Å, Billström H, Ahlquist M, Lund B. Enterococcus faecalis infection in root canals—host-derived or exogenous source? Lett Appl Microbiol 2011: 52: 109–115.
167. Nair PNR, Sjögren U, Krey G, Kahnberg K-E, Sundqvist G. Intraradicular bacteria and fungi in root-filled, asymptomatic human teeth with therapy-resistant periapical lesions: a long-term light and electron microscopic follow-up study. J Endod 1990: 16: 580–588.
168. Egan MW, Spratt DA, Ng YL, Lam JM, Moles DR, Gulabivala K. Prevalence of yeasts in saliva and root canals of teeth associated with apical periodontitis. Int Endod J 2002: 35: 321–329.
169. Siqueira JF Jr, Rôças IN. Polymerase chain reaction detection of Propionibacterium propionicus and Actinomyces radicidentis in primary and persistent endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2003: 96: 215–222.
170. Gulabivala K. Species Richness of Gram-Positive Coccoid Morphotypes Isolated from Untreated and Treated Root Canals of Teeth Associated with Periapical Disease [PhD Thesis]. University of London, 2004.
171. Siqueira JF Jr, Rôças IN. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral Microbiol Immunol 2003: 18: 263–265.
172. Zoletti GO, Pereira EM, Schuenck RP, Teixeira LM, Siqueira JF Jr, Dos Santos KR. Characterization of virulence factors and clonal diversity of Enterococcus faecalis isolates from treated dental root canals. Res Microbiol 2011: 162: 151–158.
173. Nair PNR, Schroeder HE. Periapical actinomycosis. J Endod 1984: 10: 567–570.
174. Happonen R-P. Periapical actinomycosis: a follow-up study of 16 surgically treated cases. Endod Dent Traumatol 1986: 2: 205–209.
175. O’Grady JF, Reade PC. Periapical actinomycosis involving Actinomyces israelii. J Endod 1988: 14: 147–149.
176. Sundqvist G, Reuterving C-O. Isolation of Actinomyces israelii from periapical lesion. J Endod 1980: 6: 602–606.
177. Schaal KP, Lee H-J. Actinomycete infections in humans—a review. Gene 1992: 115: 201–211.
178. Kalfas S, Figdor D, Sundqvist G. A new bacterial species associated with failed endodontic treatment: identification and description of Actinomyces radicidentis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001: 92: 208–214.
179. Brown JR, von Lichtenberg F. Experimental actinomycosis in mice. Arch Path 1970: 90: 391–402.
180. Behbehani MJ, Jordan HV. Comparative pathogenicity of Actinomyces species in mice. J Med Microbiol 1982: 15: 465–473.
181. Figdor D, Sjögren U, Sörlin S, Sundqvist G, Nair PNR. Pathogenicity of Actinomyces israelii and Arachnia propionica: experimental infection in guinea pigs and phagocytosis and intracellular killing by human polymorphonuclear leukocytes in vitro. Oral Microbiol Immunol 1992: 7: 129–136.
182. Sumita M, Hoshino E, Iwaku M. Experimental actinomycosis in mice induced by alginate gel particles containing Actinomyces israelii. Endod Dent Traumatol 1998: 14: 137–143.
183. Nair PNR, Brundin M, Sundqvist G, Sjögren U. Building biofilms in vital host tissues: a survival strategy of Actinomyces radicidentis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2008: 106: 595–603.