Basic Research—Biology
Preservation of Bacterial DNA by Human Dentin
Malin Brundin, DDS,* David Figdor, MDSc, FRACDS, DipEndo, PhD, FASM,‡ Anders Johansson, PhD,† and Ulf Sjögren, DDS, PhD*
Abstract
Introduction: The capacity of dentin and collagen to bind DNA and protect against spontaneous and nuclease-induced degradation was evaluated individually and by the incubation of DNA with nuclease-producing bacteria in a mixed culture. Methods: Extracted Fusobacterium nucleatum DNA was incubated with dentin shavings or collagen for 90 minutes. The DNA-bound substrates were incubated in different media (water, sera, and DNase I) for up to 3 months. Amplifiable DNA was released from dentin using EDTA, or from collagen using proteinase K, and evaluated by polymerase chain reaction (PCR). The stability of dentin-bound DNA was also assessed in a mixed culture (Parvimonas micra and Pseudoramibacter alactolyticus) containing a DNase-producing species, Pre-votella intermedia. Samples were analyzed for amplifiable DNA. Results: In water, dentin-bound DNA was recoverable by PCR at 3 months compared with no detectable DNA after 4 weeks in controls (no dentin). DNA bound to collagen was detectable by PCR after 3 months of incubation in water. In 10% human sera, amplifiable DNA was detectable at 3 months when dentin bound and in controls (no dentin). In mixed bacterial culture, dentin-bound DNA was recoverable throughout the experimental period (3 months), compared with no recoverable F. nucleatum DNA within 24 hours in controls (no dentin). Conclusions: There is a strong binding affinity between DNA and dentin, and between DNA and serum proteins or collagen. These substrates preserve DNA against natural decomposition and protect DNA from nuclease activity, factors that may confound molecular analysis of the endodontic microbiota yet favor paleomicro-biological studies of ancient DNA. (J Endod 2014;40:241–245)
Key Words
Bacterial nucleases, dentin, DNA binding affinity, DNA decomposition, DNA preservation, polymerase chain reaction
Under favorable conditions, host and microbial DNA can be preserved for a long period of time. The durability of teeth and the inherent preservation of dental pulp space integrity have made the dental pulp a target of scientific interest for the recovery of ancient DNA in the study of the etiology of septicemic diseases in ancient remains (1, 2) and forensic applications (3). The recovery of ancient DNA from dental pulps has been described in 400-year-old specimens (Yersina pestis) (4), 1000 year-old specimens (Mycobacterium tuberculosis) (5), and 4000-year-old specimens (Bartonella quintana) (6). These studies not only show the resilience of DNA but also highlight the favorable properties of the pulp canal space and surrounding dentin as a unique biological environment for the long-term preservation of DNA.
These environmental characteristics are an advantage for preserving DNA for paleomicrobiological studies; yet, it can be a confounding factor when applying molecular analysis to the study of the endodontic microbiota. Bacterial DNA recovered from the root canal may derive from long-dead cells reflecting the infection history, but it may have limited or no relevance for current disease.
One factor affecting DNA degradation is how cells die. If cells die with their cell membranes predominantly intact, the DNA can be protected from degradative factors and can be recovered and amplified using the polymerase chain reaction (PCR) technique. In vitro experiments using dead Enterococccus faecalis cells killed by heating to minimize cell lysis have shown that amplifiable DNA is recoverable 1 year after cell death (7), and experiments using dead E. faecalis cells inoculated in ex vivo teeth have shown recoverable DNA 2 years after cell death (8). If the bacterial cell wall disintegrates, DNA is released into the adjacent environment (Brundin, Figdor, Sundqvist, Sjögren, manuscript submitted), and the free DNA is subject to decomposition by a variety of spontaneous, ambient, and enzymatic processes (8, 9). Nucleases rapidly degrade free DNA (8).
Some environmental conditions protect DNA from spontaneous or enzymatic degradation. Hydroxyapatite, the main constituent of dentin, has a specific binding affinity for DNA (10, 11), and the resultant DNA-hydroxyapatite complex confers resistance to spontaneous degradation and protects against breakdown by nucleases (11). Collagen, the principal organic component of dentin (12), is known to bind DNA (13). Although the capability of bone (14, 15) and hydroxyapatite (11) to preserve DNA is known, the DNA-binding and protective capability of dentin and its collagen component is a largely unexplored area. Information regarding the DNA preservation capability of these structures is of importance for both endodontic and anthropologic research.
The purpose of this study was to assess the influence of dentin and collagen on the preservation of DNA from spontaneous decay. A further subject of interest was the influence of dentin on DNA preservation when DNA was incubated with nuclease-producing bacteria in a mixed culture in vitro.
Materials and Methods
Preparation of Dentin Shavings
Dentin shavings were collected from 3 intact formerly vital teeth. Teeth were stored in sterile saline and processed within 12 hours after extraction. After cleaning the teeth with sterile saline, the teeth were embedded in plaster to the cementoenamel junction. A rubber dam was applied, and the tooth crowns were carefully disinfected with H2O2 (30%, 1 minute) followed by iodine tincture (5%, 1 minute). The cervical area at the rubber dam–tooth interface was sealed with glue (Cervitec; Ivoclar Vivadent, Schaan, Lichtenstein). Disinfection was completed with a final iodine tincture wash. A conventional access cavity was prepared, and the pulp tissue was removed with Hedstrom files. Canals were filled with ultrapure water (Sigma-Aldrich, St Louis, MO), and dentin shavings were created by mechanically preparing the canal walls. The shavings were collected by aspirating the canal contents with a syringe and transferring them to a sterile Eppendorf tube. After washing (×3, ultrapure water), the dentin shavings were dried (37°C, 12 hours) and weighed. A suspension (0.1 mg dentin/μL) was prepared by suspending the shavings in ultrapure water. Ultrasonic activation was applied to disaggregate shavings.
As a sterility control, an aliquot of the dentin shavings suspension (35 μL) was plated onto Fastidious Anaerobe Agar (FAA; Laboratory M, Bury, UK) with vitamin Κ (10 mg/L) for aerobic and anaerobic incubation (37°C, 7 days). Dentin shavings (3 mg) were also treated with EDTA, processed for amplification (PCR), and analyzed for the detection of Fusobacterium nucleatum DNA (described later).
Preparation of Bacterial DNA
F. nucleatum (National Collection of Type Cultures 10652) was grown on FAA in an anaerobic box for 4 days. The cells were harvested from plates and suspended in phosphate buffered saline. DNA was extracted from cell pellets with a bacterial genomic DNA kit according to the manufacturer’s protocol (Gene Elute, Sigma-Aldrich) and eluted in ultrapure water (200 μL; Sigma-Aldrich). For all the experiments described later, DNA was prepared to a concentration of 44 ng/μL (154 ng in 3.5 μL ultrapure water).
Dentin-DNA Binding, Release, and Analysis by PCR
F. nucleatum DNA (154 ng) was added to the aqueous dentin preparations (3 mg dentin, total preparation = 100 μL) and incubated with slow agitation (22°C, 90 minutes). The DNA-coated dentin was then washed (× 3, ultrapure water), and EDTA (80 μL, 250 mmol/L, 15 minutes) was added to release DNA from dentin. The EDTA-containing supernatant was cleaned with the Wizard DNA Clean-Up System (Promega, Madison, WI) before PCR (11). For controls, identical processing was conducted for tubes containing DNA (no dentin) and tubes containing dentin (no DNA).
PCR amplifications were prepared in a 25-μL final reaction volume with 2 μL total DNA template, 12.5 pmol each primer, 200 μmοl/L dNTPs (Qiagen, Hilden, Germany), 2.5 μL 10 × PCR buffer (Qiagen) containing 1.5 mmol/L MgCl2, 0.6 U HotStarTaq DNA polymerase (Qiagen), and ultrapure water (Sigma-Aldrich) to make up the final reaction volume. Primers used were designed to amplify a 360–base pair amplicon from the F. nucleatum 16S ribosomal RNA gene (16). Amplification products (8 μL, 20 cycles) were analyzed by gel electrophoresis (1.5% agarose in Tris-borate-EDTA buffer; Gibco, Invitrogen, Grand Island, NY), stained with ethidium bromide (1 μg/mL, Sigma-Aldrich), and observed under ultraviolet light. A 100–base pair DNA ladder digest (Invitrogen, Fredrick, MD) served as a molecular weight marker. All experiments (including those described later) were repeated at least in triplicate.
Degradation of Dentin-bound DNA in Water and Sera
The degradation of dentin-bound DNA was assessed by incubating DNA (154 ng) bound to dentin (3 mg) in ultrapure water (100 μL) at 37°C for up to 3 months. The effect of serum on DNA degradation was evaluated by incubating dentin (3 mg) with DNA (154 ng, 100 μL) followed by agitation (90 minutes) and washing (×3, ultrapure water) of the dentin preparation. Sera from 4 healthy human adults working at the Department of Odontology, Umeå University, Umeå, Sweden, were pooled, inactivated (56°C, 30 minutes), and then added to the dentin preparations. Samples were analyzed at 2, 4, and 12 weeks. At these intervals, the dentin was washed (×3, ultrapure water) and incubated with EDTA (80 μL, 250 mmol/L, 15 minutes). Samples treated with EDTA were cleaned, processed by PCR (20 cycles), and analyzed as described previously. Controls were tubes containing DNA without dentin.
Dentin-bound DNA in an la Vitro Bacterial Community
The stability of dentin-bound DNA was assessed in a bacterial consortium containing DNase-producing species. To confirm bacterial survival over the observation period Parvimonas micra (Virginia Polytechnic Institute and State University 5464), Pseudoramibacter alactolyticus (Culture Collection, University of Gothenburg 52346), and Prevotella intermedia (American Type Culture Collection 25611) were grown individually for 4 days on FAA in an anaerobic box. The cells were harvested and suspended to an optical density of 1–2 in 50% human serum in phosphate buffered saline. Aliquots of the 4 strains were pooled (total volume = 4 mL) and incubated in an anaerobic box for 28 days. At predetermined intervals, samples (2 × 100 μL) were collected and analyzed by cultivation and for amplifiable DNA, respectively. Initial findings showed that the DNase producing P. intermedia survived for 2 weeks only. Therefore, in subsequent experiments, a fresh suspension of P. intermedia (106 cells in 10 μL) was added every other week over the experimental period (3 months).
F. nucleatum DNA (154 ng) bound to dentin (3 mg) was added to 100 μL of the bacterial suspension (P. micra, P. alactolyticus, and P. intermedia suspended and pooled in 50% human serum as described earlier) and incubated anaerobically for 3 months. For controls, extracted F. nucleatum DNA (without dentin) and dentin (no DNA) were individually incubated in corresponding bacterial suspensions. At predetermined intervals, samples were collected and analyzed for amplifiable DNA by PCR (30–35 cycles).
Effect of DNase on Dentin-bound DNA
The effect of nucleases on dentin-bound DNA was evaluated by binding DNA (154 ng) to dentin (3 mg) as described earlier. After binding, DNase I (1 U; Invitrogen, Carlsbad, CA) was added, and tubes were incubated with gentle agitation (22°C, 30 minutes). The dentin was washed (× 3, ultrapure water), EDTA (80 μL, 250 mmol/L) was added to release the bound DNA, and the tubes were kept on slow rotation (22°C, 15 minutes). EDTA was cleaned from samples with the Wizard DNA Clean-Up System before PCR (30 cycles), and amplification products were analyzed by gel electrophoresis. Positive controls were tubes with dentin and DNA but untreated with DNase I. Negative controls were DNA in water incubated with DNase I (without dentin).
Interaction of DNase with Dentin
The possibility of nuclease binding to dentin was assessed by incubating DNase I (0.5 U) with dentin (3 mg) in water (100 μL) with agitation (30 minutes). For analysis of DNase activity, supernatant was withdrawn (100 μL), and DNA (154 ng) was added and incubated (22°C) for 30 minutes. The DNase reaction was stopped by adding EDTA to a final concentration of 2.5 mmol/L. As a control, DNA (154 ng) was added to supernatant (100 μL) taken from the incubation of dentin not treated with DNase I. All samples were processed by PCR (30 cycles), and products were analyzed by gel electrophoresis.
Collagen-DNA Interaction
Dentin contains about 18% collagen (12), which may bind DNA. The effect of DNA binding to collagen was assessed in experiments identical to those described previously (but substituting collagen for dentin) to assess degradation in water and the effect of DNase I on collagen-bound DNA. Collagen type I from bovine Achilles tendon (2 mg; Serva, Heidelberg, Germany) was incubated in water (200 μL, Sigma-Aldrich) overnight. After washing (× 3, ultrapure water), experiments were performed as described earlier for up to 3 months. DNA was released from the collagen by the addition of proteinase K (50 μL, 100 mg/mL; 55°C, 3 hours; Sigma-Aldrich). Samples (2 μL) were processed by PCR (30–35 cycles) and analyzed by gel electrophoresis as described earlier.
Results
The sterility controls of dentin shavings revealed no viable bacteria and no detectable F. nucleatum DNA.
Dentin-DNA and Collagen-DNA Binding
DNA bound to dentin was released by the addition of EDTA. After cleanup, analysis of the supernatant revealed amplifiable DNA in all samples. Collagen-bound DNA was released by the addition of proteinase K, and analysis by PCR of the supernatant showed recoverable DNA in all samples. No DNA was detectable in controls containing dentin or collagen without added DNA.
Degradation of Dentin- and Collagen-bound DNA in Water and Dentin-bound DNA in Sera
In water, DNA was detectable by PCR only up to 4 weeks. In contrast, DNA bound to dentin was recoverable by PCR at 3 months (Fig. 1). DNA bound to collagen was just detectable (as a faint band) by PCR after 3 months of incubation (Fig. 1). In 10% human sera, DNA was detectable at 3 months. Similarly, DNA bound to dentin was recoverable at 3 months after incubation in 10% sera (Fig. 1).
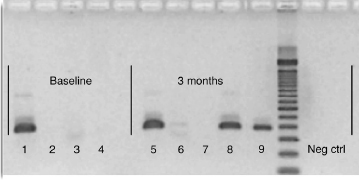
Figure 1. Recovery by PCR of F. nucleatum DNA after incubation in water or sera for 3 months. Dentin-bound, collagen-bound, and unbound DNA was released from dentin or collagen by adding EDTA or proteinase K, respectively, and assessed by gel electrophoresis of amplified DNA product (25 cycles). Baseline data: 1, DNA; 2, dentin; 3, collagen; 4, 10% sera in water. Experimental data after 3 months: 5, dentin-bound DNA in water; 6, collagen-bound DNA in water; 7, free (no dentin or collagen) DNA in water; 8, dentin-bound DNA in 10% sera; 9, free (no dentin or collagen) DNA in 10% sera. Negative control: DNA template was replaced with ultrapure water.
Dentin-bound DNA in an In Vitro Bacterial Community
For P. alactolyticus and P. micra, the initial cell density was 108 colony-forming unit/mL, and both species survived in 50% pooled human sera for 28 days. There was a gradual decline in cell density during the observation period. Amplifiable DNA for P. alactolyticus and P. micra was detectable over the whole test period (data not shown). For P. intermedia, the initial cell density was 106 colony-forming unit/mL, and it survived for 14 days. Amplifiable DNA was recoverable during the experimental period (28 days, data not shown).
When F. nucleatum DNA was inoculated into the mixed bacterial culture containing a DNase producer (P. intermedia), there was no recoverable F. nucleatum DNA within 24 hours (Fig. 2). In contrast, amplifiable DNA was recoverable throughout the whole experimental period (3 months) when it was dentin bound and inoculated into the same mixed bacterial culture (Fig. 2). When dentin-bound DNA was incubated in the same bacterial culture to which fresh P. intermedia was added every other week, DNA was recoverable at the end of the 3-month observation period (Fig. 2).
Effect of Nucleases on Dentin- and Collagen-bound DNA
Dentin-bound DNA was incompletely degraded by DNase I, as shown by detectable DNA after EDTA treatment and cleanup of the supernatant. Similarly, DNA bound to collagen was also detectable after exposure to DNase I. In the control (ie, DNA incubated with DNase I in water [30 minutes]), there was no PCR-detectable DNA.
Interaction of DNase with Dentin and Collagen
DNase activity was evaluated after the incubation of DNase I with dentin and collagen, respectively, by the addition of DNA to their supernatants. Nonrecovery of DNA would indicate unimpaired DNase activity. After treatment, DNA was still detectable, which implies that activity was inhibited after the incubation of DNase with dentin or collagen. In the control (no dentin or collagen), DNA was degraded within 30 minutes as shown by the nonrecovery of DNA.
Discussion
Bone and teeth are generally recognized as favorable sites for preservation and potential recovery of ancient DNA (17). Apart from the physical protection afforded by these hard tissues, the main inorganic component, hydroxyapatite, has a strong binding affinity for DNA (10, 11). It has recently been shown that there is a significant reduction in spontaneous degradation when DNA is bound to hydroxyapatite (11). The present study assessed dentin as a substrate for DNA preservation and the results show that dentin-bound DNA is conserved from spontaneous decomposition when compared with free DNA. Binding to other biomolecules (ie, collagen and serum) was also shown to have a preservative effect against spontaneous decomposition.
When microbes die with their cell walls predominantly intact, it has been shown experimentally that cell-bound bacterial DNA is very stable (7, 8). If the cell wall breaks open, DNA is released (Brundin, Figdor, Sundqvist, Sjögren, manuscript submitted) and is thereby exposed to environmental factors that may accelerate or delay decay (8). Water and nucleases are environmental factors that can induce rapid decomposition of naked DNA molecules within hours to weeks (8, 9).
DNA may remain detectable for tens or hundreds of years (4, 5, 18), which implies that there are favorable ambient conditions that can stabilize the DNA molecule or protect it from degradative factors. DNA can bind to minerals in sand and clay (19, 20) and to hydroxyapatite (10, 11). Laboratory studies show a strong binding affinity of hydroxyapatite for DNA (10, 11). The DNA-mineral complex stabilizes the DNA by changing the charge of the DNA molecule (21). The present study showed that dentin-bound DNA incubated in water was recoverable for 3 months compared with undetectable DNA after 4 weeks without dentin. This finding may seem unsurprising because hydroxyapatite is the main component of dentin (22); yet, dentin also contains a significant quantity of proteins (23), predominantly collagen type I (24), which may influence binding to DNA. Additionally, serum may enter the root canal by diffusion through the apical foramen or via the vessels and bind free DNA (25). Therefore, DNA binding to collagen and serum were assessed as separate biological components.
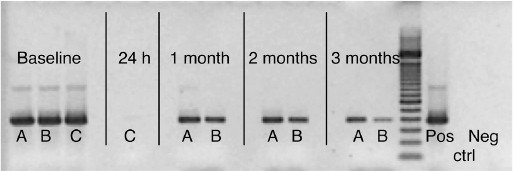
Figure 2. Recovery by PCR of F. nucleatum DNA after coculture in a bacterial consortium (P. micra, P. alactolyticus and P. intermedia in 50% sera) with and without dentin. Analysis by gel electrophoresis of amplified DNA product. Data collected at baseline; 24 hours; and 1, 2, and 3 months. A, Dentin-bound DNA incubated in bacterial mix; B, dentin-bound DNA incubated in bacterial mix with the addition of P. intermedia every other week; C, free (no dentin or collagen) DNA incubated in bacterial mix. Controls: positive amplification control (Pos) and negative control; DNA template was replaced with ultrapure water (Neg).
DNA was shown to bind to collagen, which is consistent with previous reports (13, 26), and the DNA was recoverable after collagen removal by the addition of proteinase K. Collagen-bound DNA was barely detectable after 3 months in water, which suggests a weak binding affinity and a limited protective effect on DNA preservation. Binding to serum had a preservative effect because DNA exposed to sera was recoverable after 3 months. DNA binds to serum proteins (25), in particular to albumin, the most abundant protein, binding DNA between the protein and the G-C bases and backbone PO2 group of the DNA molecule, resulting in 2 different albumin-DNA complexes with different bonding strengths (25). In a supplementary experiment using collagen-bound DNA incubated in serum for 3 months, there was limited amplifiable DNA associated with the collagen component; yet, the serum supernatant contained ample recoverable DNA (data not shown). This finding implies a variable binding affinity between different tissue proteins and that serum proteins bind more strongly to DNA than collagen. Human sera also contain some DNases (27); yet, these low concentrations (3–23.9 ng/mL) evidently have little impact on DNA degradation when bound to dentin.
In the root canal, it has been suggested that infection and, in particular, the production of innate microbial DNases may contribute to DNA decomposition (28). Previous in vitro experiments have shown that free E. faecalis DNA is rapidly degraded after incubation in culture with P. intermedia; yet, the effect was not observed for cell-bound E. faecalis DNA (8). These interactions are undoubtedly complex; yet, the results showing the protective effect of both hydroxyapatite (11) and dentin (this study) together with paleomicrobiological data (4, 6) would suggest that these minerals confer a protective effect against microbial nucleases in root canal infections. Therefore, the effect of dentin on protecting DNA from nuclease-producing bacteria was assessed by incubating DNA in a mixed culture containing P. micra and P. alactolyticus together with P. intermedia, selected for its potent nuclease capability (8, 29, 30).
In mixed bacterial cultures containing dentin, amplifiable DNA was recoverable at 3 months compared with controls (no dentin) with no detectable DNA after 24 hours, which shows a pronounced protective effect of dentin. In some experiments, fresh P. intermedia culture was added every second week to ensure that DNase-producing activity was maintained throughout the observation period, and the findings were the same; amplifiable F. nucleatum DNA was detected at 3 months. These results show the significance of dentin in both preservation of DNA and mitigating the degradative activity of nucleases in bacterial infection.
The contribution of individual components was assessed in more detail by separately incubating dentin- and collagen-bound DNA with DNase I. Amplifiable DNA was recovered after the incubation of dentin-bound DNA and collagen-bound DNA with DNase I, which shows that the binding of DNA to both substrates individually affords protection from degradation by DNase. These findings are consistent with that seen for hydroxyapatite, which also protects DNA from nuclease activity (11).
The mechanisms for protection against nuclease degradation may include dentin or collagen binding to DNA, making the molecule inaccessible to enzymatic activity, adsorption of the DNase itself to biological substrates, or a combination of both. Hydroxyapatite has been shown to bind DNase I, which contributes to the effectiveness of hydroxyapatite in preserving DNA against nuclease activity (11). In this study, pretreatment of dentin and collagen with DNase I inhibited subsequent nuclease activity. Taken together, the data show that binding to dentin reduces the degradative efficacy of nuclease-producing bacteria and that DNase activity is inhibited individually by hydroxyapatite, dentin, and collagen.
Extracellular DNA is derived from decomposing or disrupted cells or via excretion from live cells (31). Recent studies have shown that excreted extracellular DNA is a natural part of biofilm development and growth (32–34). In the infected root canal, intimate contact of such extracellular DNA on dentin would significantly enhance the preservation of bacterial DNA. It is reasonable to assume that DNA so adsorbed to dentin would then be recoverable a long time thereafter, a factor that should be taken into account when processing samples for molecular analysis.
The results presented herein show a strong binding affinity between DNA and dentin, which is consistent in many ways with previous observations for hydroxyapatite and DNA (10, 11). The resultant dentin-DNA complex preserves DNA against natural decomposition in water and protects DNA against nuclease activity in mixed bacterial culture. These findings help explain why ancient DNA is so well preserved within the pulp space of teeth in centuries-old human remains (1, 3, 6). The binding affinity is variable between different biological substrates; dentin appears to possess a greater affinity than serum proteins or collagen for DNA. The findings also have implications for research in endodontic microbiology; the limitations must be considered, and special measures may be necessary for root canal samples processed using PCR.
Acknowledgments
The authors thank Mrs Chrissie Roth for invaluable technical assistance and Professor Göran Sundqvist for critical review of the manuscript.
Supported by the County of Västerbotten, Public Dental Health, and the Australian Dental Research Foundation Inc.
The authors deny any conflicts of interest related to this study.
References
- Raoult D, Aboudharam G, Crubezy E, et al. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of medieval black death. Proc Natl Acad Sci U S A 2000;97:12800–3.
- Drancourt M, Raoult D. Palaeomicrobiology: current issues and perspectives. Nat Rev Microbiol 2005;3:23–35.
- Potsch L, Meyer U, Rothschild S, et al. Application of DNA techniques for identification using human dental pulp as a source of DNA. Int J Legal Med 1992;105:139–43.
- Drancourt M, Aboudharam G, Signoli M, et al. Detection of 400-year-old Yersinia pestis DNA in human dental pulp: an approach to the diagnosis of ancient septicemia. Proc Natl Acad Sci U S A 1998;95:12637–40.
- Donoghue HD, Marcsik A, Matheson C, et al. Co-infection of Mycobacterium tuberculosis and Mycobacterium leprae in human archaeological samples: a possible explanation for the historical decline of leprosy. Proc Biol Sci 2005;272:389–94.
- Drancourt M, Tran-Hung L, Courtin J, et al. Bartonella quintana in a 4000-year-old human tooth. J Infect Dis 2005;191:607–11.
- Young G, Turner S, Davies JK, et al. Bacterial DNA persists for extended periods after cell death. J Endod 2007;33:1417–20.
- Brundin M, Figdor D, Roth C, et al. Persistence of dead-cell bacterial DNA in ex vivo root canals and influence of nucleases on DNA decay in vitro. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2010;110:789–94.
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993;362: 709–15.
- Bernardi G. Chromatography of nucleic acids on hydroxyapatite. Nature 1965;206: 779–83.
- Brundin M, Figdor D, Sundqvist G, et al. DNA binding to hydroxyapatite: a potential mechanism for preservation of microbial DNA. J Endod 2013;39:211–6.
- Stack MV. The chemical nature of the organic matrix of bone, dentin, and enamel. Ann Ν Υ Acad Sci 1955;60:585–95.
- Rosenberg AM, Hunt DW, Petty RE. The binding of DNA to native type Π collagen. J Rheumatol 1983;10:925–9.
- Pruvost M, Schwarz R, Correia VB, et al. Freshly excavated fossil bones are best for amplification of ancient DNA. Proc Natl Acad Sci U S A 2007;104:739–44.
- Götherström A, Collins M, Angerbjörn A, et al. Bone preservation and DNA amplification. Archaeometry 2002;44:395–404.
- Conrads G, Gharbia SE, Gulabivala K, et al. The use of a 16s rDNA directed PCR for the detection of endodontopathogenic bacteria. J Endod 1997;23:433–8.
- Lassen C, Hummel S, Herrmann B. Comparison of DNA extraction and amplification from ancient human bone and mummified soft tissue. Int J Legal Med 1994;107: 152–5.
- Wandeler P, Smith S, Morin PA, et al. Patterns of nuclear DNA degeneration over time—a case study in historic teeth samples. Mol Ecol 2003;12:1087–93.
- Goring CAI, Bartholomew WV. Adsorption of mononucleotides, nucleic acids, and nucleoproteins by clays. Soil Sci 1952;74:149–64.
- Lorenz MG, Wackernagel W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl Environ Microbiol 1987;53:2948–52.
- Khanna M, Stotzky G. Transformation of Bacillus subtilis by DNA bound on mont-morillonite and effect of DNase on the transforming ability of bound DNA. Appl Environ Microbiol 1992;58:1930–9.
- Marshall GW Jr. Dentin: microstructure and characterization. Quintessence Int 1993;24:606–17.
- Jagr M, Eckhardt A, Pataridis S, et al. Comprehensive proteomic analysis of human dentin. Eur J Oral Sci 2012;120:259–68.
- Park ES, Cho HS, Kwon TG, et al. Proteomics analysis of human dentin reveals distinct protein expression profiles. J Proteome Res 2009;8:1338–46.
- Malonga H, Neault JF, Arakawa H, et al. DNA interaction with human serum albumin studied by affinity capillary electrophoresis and FTIR spectroscopy. DNA Cell Biol 2006;25:63–8.
- Svintradze DV, Mrevlishvili GM, Metreveli Ν, et al. Collagen-DNA complex. Bio-macromolecules 2008;9:21–8.
- Miyauchi K, Ogawa M, Murata A, et al. Serum deoxyribonuclease I determined by a radioimmunoassay and an enzymatic assay in malignant diseases. Clin Chim Acta 1989;184:115–9.
- Siqueira JF Jr. On the issue of uncultivated bacteria and dead cell detection by molecular methods: reply to Dr. Nair’s commentary. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2008;105:5–8. author reply 8–10.
- Leduc A, Grenier D, Mayrand D. Outer membrane-associated deoxyribonuclease activity of Porphyromonas gingivalis. Anaerobe 1995;1:129–34.
- Porschen RK, Sonntag S. Extracellular deoxyribonuclease production by anaerobic bacteria. Appl Microbiol 1974;27:1031–3.
- Nielsen KM, Johnsen PJ, Bensasson D, et al. Release and persistence of extracellular DNA in the environment. Environ Biosafety Res 2007;6:37–53.
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, et al. Extracellular DNA required for bacterial biofilm formation. Science 2002;295:1487.
- Martins M, Uppuluri P, Thomas DP, et al. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 2010;169:323–31.
- Barnes AM, Ballering KS, Leibman RS, et al. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. MBio 2012;3. e00193–12.
From the Departments of *Odontology/Endodontics and † Odontology/Molecular Periodontology, Faculty of Medicine, Umeå University, Umeå, Sweden; and ‡Department of Microbiology, Monash University, Melbourne, Australia.
Address requests for reprints to Dr Malin Brundin, Department of Odontology/Endodontics, Faculty of Medicine, Umeå University, Umeå, 90187, Sweden. E-mail address: [email protected] 0099-2399/$ - see front matter
Copyright © 2014 American Association of Endodontists. http://dx.doi.org/10.1016/j.joen.2013.08.025