Basic Research—Biology
DNA Binding to Hydroxyapatite: A Potential Mechanism for Preservation of Microbial DNA
Malin Brundin, DDS,* David Figdor, MDSc,
FRACDS, DipEndo, PhD, FASM,†
Göran Sundqvist, DDS, PhD,* and Ulf Sjögren, DDS, PhD*
Abstract
Introduction: Molecular methods are increasingly being deployed for analysis of the microbial flora in the root canal. Such methods are based on the assumption that recovered DNA is associated with the active endodontic infection, yet paleomicrobiology research is based on the recovery of ancient DNA from centuries-old tooth and bone samples, which points to considerable longevity of the DNA molecule in these tissues. The main component of dentin and bone is the mineral hydroxyapatite. This study assessed DNA binding to hydroxyapatite and whether this binding affinity stabilizes the DNA molecule in various media. Methods: DNA was extracted from Fusobacterium nucleatum and added to ceramic hydroxyapatite for 90 minutes. The DNA-bound hydroxyapatite was incubated in different media (ie, water, sera, and DNase I) for up to 3 months. At predetermined intervals, the recovery of detectable DNA was assessed by releasing the DNA from the hydroxyapatite using EDTA and evaluating the presence of DNA by gel electrophoresis and polymerase chain reaction (PCR) amplification. Results: When incubated with hydroxyapatite, nonamplified DNA was detectable after 3 months in water, sera, and DNase I. In contrast, DNA incubated in the same media (without hydroxyapatite) decomposed to levels below the detection level of PCR within 3 weeks, with the exception of DNA in sera in which PCR revealed a weak positive amplification product. Conclusions: These results confirm a specific binding affinity of hydroxyapatite for DNA. Hydroxyapatite-bound DNA is more resistant to decay and less susceptible to degradation by serum and nucleases, which may account for the long-term persistence of DNA in bone and tooth. (J Endod 2013;39:211–216)
KeyWords
Ancient DNA, DNA binding affinity, DNA decomposition, DNA preservation, hydroxyapatite, polymerase chain reaction
Modern endodontic microbiology was established using advanced anaerobic techniques for clinical recovery and culture of root canal samples (1–3). For more than a decade, molecular techniques based on the recovery of specific microbial DNA sequences that can be amplified by the polymerase chain reaction (PCR) method (4) have been increasingly used for analysis of the microbial flora (5, 6). Studies using molecular techniques have generally reported a more diverse microbial flora associated with endodontic infections than that revealed by culture methods.
The PCR method affords specificity and precision to microbial identification; however, there are limitations of the PCR method when applied to root canal samples. A principal issue is the fate of DNA from bacteria that may enter and die in the root canal and circumpulpal dentin because the PCR method does not distinguish between living and dead cells (7). Should DNA from dead cells persist, it could mean that some species identified by recovered DNA could be of minor importance or no relevance for the disease. Although cell-bound DNA can be detected by PCR methods 2 years after cell death, free DNA degrades spontaneously and by enzymatic action (8).
The binding affinity of DNA to hydroxyapatite (HA) may influence how long free DNA persists in the root canal space and may contribute to the successful recovery of ancient DNA from old teeth and bone samples (9, 10). The main component (~60%) of dentine, HA (11), stabilizes DNA and slows the rate of hydrolytic depurination (12). Mineral-adsorbed DNA is 100- to 1,000-fold more resistant to decay than free DNA (13, 14). In early work, after the technique was developed for protein separation (15), HA columns were used as an efficient binding method for sequestering nucleic acids (16).
The HA binding property may confer protection to DNA from degradation by spontaneous decay and enzymatic decomposition by ambient nucleases, yet these properties are essentially unexplored in the endodontic literature. The aim of the present study was to investigate the following:
- The ability of HA to bind extracellular DNA
- A method to release HA-bound DNA and prepare it in a form suitable for PCR
- The influence of HA binding on protecting DNA from degradation by serum, nucleases, and decay over time
Materials and Methods
Bacteria
Fusobacterium nucleatum (NCTC 10652) was grown on fastidious anaerobe agar (Laboratory M, Bury, UK) with vitamin K (10 mg/L, 37°C, 4 days) in an anaerobic box (10% H2 and 5% CO2 in N2). The cells were harvested from plates and suspended in phosphate-buffered saline solution. DNA was extracted from cell pellets according to the manufacturer’s protocol (Bacterial Genomic DNA Kit; Gene Elute, Sigma-Aldrich, St Louis, MO).
HA-DNA Binding, Release, and Analysis
Ceramic HA type II (BioRad, Hercules, CA) of 80-µm particle size (0.8–9.4 mg) was suspended in sterile water (93 µL) and incubated at room temperature overnight. Extracted F. nucleatum DNA (318 ng) was added to the aqueous HA preparations and incubated with slow agitation (22°C, 90 minutes). Controls were tubes containing DNA without HA.
After incubation, residual DNA in the supernatant was assessed by taking aliquots of supernatant (10 and 2 µL, respectively) for analysis by gel electrophoresis (1.5% agarose in Tris-borate EDTA buffer; Gibco, Invitrogen, Grand Island, NY), for staining with ethidium bromide (1 µg/mL; Sigma-Aldrich), for observation under ultraviolet light, and for PCR analysis as described later. Thereafter, the HA was washed (3×), and EDTA (80 µL, 250 mmol/L, 15 minutes) was added to release DNA. EDTA-induced release of DNA from HA was confirmed by aliquot removal (10 µL) and analysis by gel electrophoresis. A 100–base pair DNA ladder digest (Invitrogen, Fredrick, MD) served as a molecular weight marker.
The optimal conditions for DNA release by EDTA were assessed by suspending HA (1–10 mg) in sterile water (145.5 µL) and incubation overnight at room temperature. Extracted F. nucleatum DNA (205 ng) was added to the HA preparations and incubated with slow agitation (90 minutes). After incubation, HA beads were washed (3×), and EDTA (140 µL; 250 mmol/L) was added. At predetermined intervals (15 minutes–24 hours), aliquots of supernatant (10 µL) were removed and analyzed by gel electrophoresis.
A high concentration of EDTA interferes with the PCR reaction (17, 18). Therefore, for PCR, the EDTA-containing supernatant was cleaned with the Wizard DNA Clean-Up System (Promega, Madison, WI) according to the manufacturer’s protocol. Briefly, samples were added to 1 mL Wizard DNA Clean-Up Resin and transferred to a syringe barrel with a minicolumn. The bound DNA was pushed through the minicolumn with a syringe plunger. The column was then washed with 2 mL isopropanol (80%) and transferred to a microcentrifuge tube. After centrifugation (2 minutes, 10,000g), the minicolumn was transferred to a new microcentrifuge tube, and 50 µL prewarmed (65°C) Tris EDTA buffer was added. The minicolumn was centrifuged (20 seconds, 10,000g) to elute the bound DNA. Experiments with various concentrations of EDTA showed that this method allowed effective PCR reactions.
For PCR, primers were used to amplify a 360–base pair amplicon from the F. nucleatum 16S ribosomal RNA gene (5). PCR amplifications were prepared in a 25-µL final reaction volume with 2 µL total DNA template, 12.5 pmol each primer, 200 µmol/L dNTPs (Qiagen, Hilden, Germany), 2.5 µL 10× PCR buffer (Qiagen), 0.6 U HotStarTaq DNA polymerase (Qiagen), and Ultrapure water (Sigma-Aldrich) to make up the final reaction volume. Amplification products were analyzed by gel electrophoresis as described earlier. All experiments (including those described later) were repeated at least 3 times.
Effect of Nucleases on HA-bound DNA
HA binding of DNA may confer protection from degradation by nucleases. The effect was evaluated by binding DNA (318 ng) to HA (9 mg) as described previously. After binding DNA to HA, DNase I (Invitrogen, Grand Island, NY; 1 U) was added, and tubes were incubated with gentle agitation (22°C, 30 minutes). The HA was then washed (3×) with sterile water. EDTA (80 µL; 250 mmol/L) was added to release DNA from the HA, and the tubes were kept on slow rotation (22°C, 15 minutes). Supernatant was withdrawn (10 µL), and the release of DNA from HA was analyzed by gel electrophoresis. Positive controls were tubes with HA and DNA but untreated with DNase I. DNA in water incubated with DNase I (without HA) served as the negative controls. EDTA was cleaned from samples as described earlier followed by PCR, and amplification products were analyzed by gel electrophoresis.
Interaction of DNase with HA
Because DNase binding to HA could affect nuclease function, DNase activity was analyzed after incubation with HA. DNase I (0.5 U) was incubated with HA (1–20 mg) in water (100 µL) with agitation (30 minutes). For the analysis of DNase activity, supernatant was withdrawn (100 µL), and DNA (3.5 µL; 44 ng/µL) was added and incubated (22°C) for 30 minutes. The DNase reaction was stopped by adding EDTA to a final concentration of 2.5 mmol/L. An aliquot (10 µL) was withdrawn and analyzed for DNA by gel electrophoresis. As a control, DNase I (0.5 U) was added to supernatant taken from an overnight incubation of HA in water (100 µL). PCR amplification was performed (17 cycles) on all samples, and products were analyzed by gel electrophoresis.
Degradation over Time of HA-bound DNA
DNA (159 ng) was bound to HA (9 mg) as described previously. The tubes were incubated (37°C) and subsequently sampled at 2, 4, 8, and 12 weeks. At harvest, the HA was washed (3×) and incubated with EDTA (80 µL; 250 mmol/L, 15 minutes). Supernatant (10 µL) was analyzed by gel electrophoresis. Controls were tubes containing DNA but without HA treated in the same way, and 10-µL aliquots were analyzed by gel electrophoresis. Samples treated with EDTA were cleaned and analyzed by PCR as described previously.
Effect of Human Serum on Degradation over Time of HA-bound DNA
Sera from 4 healthy human adults were pooled and inactivated at 56°C for 30 minutes. HA (9 mg) was incubated with DNA (318 ng; 100 µL) with agitation (90 minutes) then washed (3 ×), and the pooled sera (10%) was added. After 3 months of incubation, the HA was washed and treated with EDTA (80 µL; 250 mmol/L, 15 minutes). An aliquot (10 µL) was withdrawn and analyzed by gel electrophoresis. The remainder of the supernatant was cleaned with the Wizard Clean-Up System followed by PCR, and amplification products were analyzed by gel electrophoresis. Controls were DNA in sera without HA.
Results
HA-DNA Binding and Release
When EDTA was added to HA-bound DNA, there was an optimal release of DNA over the first 15 minutes to 2 hours. After 4 hours, there was a reduction of measurable DNA, and by 7 hours DNA was no longer detectable in the supernatant of tubes containing 7 mg or more of HA.
After binding extracted DNA (318 ng) to ceramic HA, nonamplified DNA was not detectable in the supernatant from tubes containing ≥5.6 mg HA (Fig. 1A). With amplification by PCR (20 cycles), DNA was detectable in all samples (data not shown). DNA was released from HA by the addition of EDTA. Analysis of the supernatant revealed a dose relationship between HA mass and DNA binding; a greater HA mass bound more DNA (Fig. 1B).
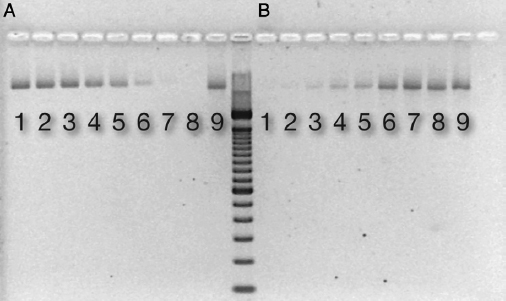
Figure 1. DNA binding to ceramic HA. An electrophoresis gel picture showing detectable DNA in supernatant after binding to increasing amounts of (A) HA and (B) after releasing DNA from HA by the addition of EDTA. Respective lanes contain the following mass of ceramic HA type II: (1) 0.8 mg, (2) 1.3 mg, (3) 1.8 mg, (4) 3.0 mg, (5) 3.5 mg, (6) 5.6 mg, (7) 7.4 mg, (8) 9.4 mg, and (9) control: H2O + DNA (318 ng).
Effect of Nucleases on HA-bound DNA
HA-bound DNA was resistant to degradation by DNase I as shown by the detection of DNA after EDTA treatment and cleanup of HA (Table 1). In contrast, there was no detectable DNA after incubation with DNase I in water (30 minutes).
Interaction of DNase with HA
There was a loss of DNA degradation activity when DNase I was incubated with HA, an effect that was dose related to the amount of HA. DNA added to treated supernatant (ie, DNase I incubated with HA in water) was still detectable in both the nonamplified and amplified (PCR, 20 cycles) form if the amount of HA was ∵5 mg (Fig. 2A and B). In the control (no HA), DNA was degraded in 30 minutes as shown by the nonrecovery of DNA (Fig. 2A and B).
Degradation over Time of HA-bound DNA
In water, DNA decomposed within 2 weeks to below the detection level of gel electrophoresis (nonamplified DNA) although it was detectable by PCR at 4 weeks (but not at 8 weeks). After binding to HA for 90 minutes, DNA was recoverable in nonamplified form and by PCR for >3 months (Table 1).
Influence of Sera on HA-bound DNA
In 10% human sera, DNA decayed within 2 weeks to below the detection level of gel electrophoresis (nonamplified DNA). With PCR, DNA was detectable (faint bands) for 3 months. By contrast, DNA bound to HA did not degrade after 3 months in 10% sera as shown by recoverable nonamplified DNA (Table 1).
Discussion
Molecular-based methods have become increasingly favored for analysis of the composition of microbial DNA within samples taken from infected root canals. If the DNA is derived from living microorganisms in a noncontaminated root canal sample, molecular analysis should reflect the composition of the active bacterial flora. Yet, there are indications that long after cell death DNA and DNA fragments may linger for centuries in bone and teeth, which can be detected by the amplification of DNA using PCR (9, 19). If the same applies to infected root canals (ie, that DNA from dead bacteria persists and is recoverable by PCR), the findings in some cases may be a distortion of the true connection between species so identified and the active endodontic infection. A binding affinity between HA and DNA may account for the persistence of ancient DNA in teeth and bone (9, 10). This study assessed the binding of F. nucleatum DNA to HA and whether the mineral-DNA complex resists decay. The results show that, compared with unbound DNA, HA-bound DNA was much more resistant to spontaneous decay and was less susceptible to degradation by serum and nucleases.
It has been well established that HA binds DNA (16, 19), and the results herein confirmed this binding affinity. Binding occurred in a dose-related way, so that with a greater mass of HA (>5.6 mg HA and 318 ng DNA) there was no detectable DNA left in the supernatant. Binding to HA had a stabilizing effect on spontaneous DNA decay, as shown by recoverable DNA after >3 months in water. In contrast, non-bound DNA in water was undetectable after 4 weeks of incubation.
Table 1. Recovery of Free and HA-bound DNA after Incubation in Water, with DNase, and in 10% Human Sera
| Spontaneous degradation* | Effect of nucleases† | Influence of sera‡ | ||||
| Free DNA | HA-bound DNA | Free DNA | HA-bound DNA | Free DNA | HA-bound DNA | |
| Nonamplified DNA | – | + | – | + | – | + |
| DNA amplified by PCR | – | + | – | + | +§ | + |
Results from triplicate experiments.
*DNA incubated in water for 3 months.
†DNA incubated in DNase I for 30 minutes.
‡DNA incubated in sera for 3 months.
§Weak band.
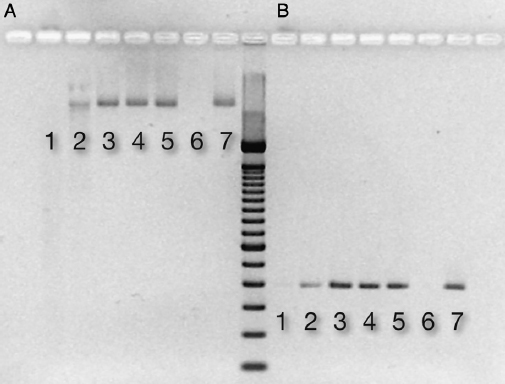
Figure 2. Nuclease activity after DNase incubation with ceramic HA. After incubation, nuclease activity in the supernatant was assessed by adding DNA for 30 minutes. The electrophoresis gel picture shows detectable DNA in (A) the nonamplified form and (B) after PCR amplification. Respective lanes contain the following mass of ceramic HA type II: (1) 1 mg, (2) 5 mg, (3) 10 mg, (4) 15 mg, (5) 20 mg, (6) control: DNase (0.5 U) + DNA (150 ng), and (7) control: DNA (150 ng) in water.
Ion attraction is the likely mechanism for the binding affinity of DNA to mineral surfaces. The binding capacity is influenced by the presence of cations (13), and bivalent cations result in a 100-fold higher adsorption compared with monovalent ions (14). The cations may act as a bridge between the negatively charged ion groups on the mineral surface and the DNA (20, 21). Furthermore, a low pH enhances the adherence capacity (13, 22–24) as protonation of the negatively charged DNA groups inhibit the repelling forces to the negatively charged groups on the mineral surface (24) in favor of molecular adherence.
The chelating agent, EDTA, is commonly used for the retrieval of DNA from tooth and bone samples (10, 25, 26). Although EDTA is efficient for this application, its presence in the mixture inhibits the PCR reaction. Therefore, we tested a variety of decontamination approaches and found that a proprietary system (Wizard DNA Clean-Up System) was effective for the removal of EDTA, resulting in DNA release and no inhibition of the PCR reaction. The release of DNA from HA was confirmed by treatment with a weak (250 mmol/L) EDTA solution. A maximum release of DNA was found after 15 minutes, and extending the EDTA treatment time beyond 4 hours resulted in a lower yield, probably because of saturation of the EDTA-calcium ion complex resulting in a rebinding of DNA to HA.
Apart from spontaneous decay, local environmental factors significantly influence DNA longevity or degradation (8, 12). The infected root canal likely contains small amounts of serum that perfuses into the space from the apical tissue. Human serum contains a low concentration (3–23.9 ng/mL) of DNase (27, 28). It has been previously shown that DNA incubated in pooled human sera decomposed to below the detection limit within 24 hours (8). Using gel electrophoresis, this study confirmed those findings for nonamplified DNA. With a higher level of sensitivity provided by amplification, there was faint PCR-detectable DNA after 3 months of incubation in serum (Table 1). By contrast, there was a strong stabilizing effect on DNA when it was HA bound as shown by the detection of DNA in both nonamplified form and by PCR after 3 months of incubation in pooled human sera.
The observation that there were low levels of detectable DNA after serum exposure points to competing factors in serum that favor preservation as well as degradation of DNA. The binding of DNA to serum proteins may account for some protection because serum proteins have been shown to bind DNA (29, 30). It is possible that most of the DNA decomposes naturally or by host nucleases, but small amounts of DNA that bind to serum proteins may escape decomposition.
In the root canal milieu, microbial DNases may actively degrade microbial DNA, but there has been limited analysis of the contribution of HA to this process. The role of HA was assessed by the incubation of DNase with HA-bound DNA, and this showed that recovery of DNA was possible after exposure to DNase. In contrast, without HA, there was enzymatic breakdown of DNA by DNase within 30 minutes (Table 1). This implies a protective effect of HA against nuclease activity, which could be caused by binding of the DNA to HA, making the molecule inaccessible to the enzymatic activity of DNase. An alternative explanation is a simultaneous adsorption of DNase itself directly to HA.
It has been reported that minerals have DNase adsorption capacity (31) and that DNase loses its degrading capacity when it binds to clay particles. Therefore, DNase activity was assessed with and without pre-exposure to HA. When DNase was pre-exposed to HA, there was a reduction in the nuclease activity, and the effect was dose related; increased proportions of HA resulted in lower DNase activity. Thus, we showed that the mechanism for protection against enzymatic activity is a combination of binding of both DNase and binding of the DNA molecules to HA. These findings are consistent with a study that showed the protection of plasmid DNA from nuclease activity by adsorption of DNase to various clay minerals (32).
In previous studies, cell-bound DNA was shown to be very stable and was detectable by PCR years after cell death in vitro (33) and in root canals ex vivo (8). However, free (ie, non–cell bound) DNA decomposed spontaneously after a short time period (8), which highlights the role that the form of DNA plays in its longevity. The findings reported here add further information to the complex picture and show the importance of HA in protecting DNA from natural decay and degradation by serum and nucleases. It supports the idea that binding to HA may contribute to the preservation of ancient microbial DNA in teeth and bone.
There are implications of these findings for PCR-based studies of the endodontic microbial flora. The intimate contact of extracellular DNA, whether from a living microbial biofilm (34, 35) or from dead cells, affords the possibility of DNA adsorption to HA. The protection provided by HA binding raises the possibility that DNA from past infection may be bound by HA and preserved, to be picked up later and potentially confound molecular analysis of the root canal sample. For example, some studies have reported a higher prevalence of the readily cultivable Enterococcus faecalis by PCR (36, 37) compared with culture (38, 39). The observation has apparently been difficult to explain, other than by a presumed (yet not proven) higher sensitivity of PCR (36, 37). Notwithstanding other considerations, the findings from the present study could account for the recovery of such DNA (bound to the HA constituent) from human root canals. Further factors that may result in misleading positive or negative results are the observations that a weak EDTA solution is effective in releasing DNA bound to HA and that EDTA carryover into the PCR mixture may inhibit the reaction. It is worth noting that these findings apply to HA in vitro, and whether the binding affinity also applies to dentin is an issue that awaits future clarification. Nevertheless, the results taken as a whole show the complex interaction between many factors that influence DNA degradation or preservation in dentin.
There is little doubt about the value of PCR-based methods for the identification of microbial DNA, yet there remain some important questions about how it is implemented in the context of sampling the infected root canal. The fate of DNA, expressed from dead bacteria and left in the root canal, is a potential confounder in endodontic molecular microbiology studies and has been a contentious issue in the endodontic literature (40–42). Whether DNA persists in the root canal depends on its form (free or cell bound); the root canal milieu; and whether the DNA will be exposed to factors that enhance or reduce decay such as temperature, moisture, pH, and oxidative agents (12, 43). This study shows that HA, the main constituent of dentin and bone, has a strong stabilizing role in protecting DNA from natural decay and degradation by ambient environmental factors.
Acknowledgments
The authors thank Mrs Chrissie Roth for invaluable technical assistance and Dr Anders Johansson for critical review of the manuscript.
The authors deny any conflicts of interest related to this study.
References
1. Kantz WE, Henry CA. Isolation and classification of anaerobic bacteria from intact pulp chambers of non-vital teeth in man. Arch Oral Biol 1974;19:91–6.
2. Wittgow WC Jr, Sabiston CB Jr. Microorganisms from pulpal chambers of intact teeth with necrotic pulps. J Endod 1975;1:168–71.
3. Sundqvist G. Bacteriological studies of necrotic dental pulps [odontological dissertation]. Umeå, Sweden: Umeå University; 1976.
4. Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 1987;155:335–50.
5. Conrads G, Gharbia SE, Gulabivala K, et al. The use of a 16s rDNA directed PCR for the detection of endodontopathogenic bacteria. J Endod 1997;23:433–8.
6. Siqueira JF Jr, Rôças IN. PCR methodology as a valuable tool for identification of endodontic pathogens. J Dent 2003;31:333–9.
7. Josephson KL, Gerba CP, Pepper IL. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl Environ Microbiol 1993;59:3513–5.
8. Brundin M, Figdor D, Roth C, et al. Persistence of dead-cell bacterial DNA in ex vivo root canals and influence of nucleases on DNA decay in vitro. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2010;110:789–94.
9. Drancourt M, Aboudharam G, Signoli M, et al. Detection of 400-year-old Yersinia pestis DNA in human dental pulp: an approach to the diagnosis of ancient septicemia. Proc Natl Acad Sci U S A 1998;95:12637–40.
10. Wandeler P, Smith S, Morin PA, et al. Patterns of nuclear DNA degeneration over time—a case study in historic teeth samples. Mol Ecol 2003;12:1087–93.
11. Bowes JH, Murray MM. The chemical composition of teeth: the composition of human enamel and dentine. Biochem J 1935;29:2721–7.
12. Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993;362: 709–15.
13. Lorenz MG, Wackernagel W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl Environ Microbiol 1987;53:2948–52.
14. Romanowski G, Lorenz MG, Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol 1991;57:1057–61.
15. Tiselius A, Hjertén S, Levin Ö. Protein chromatography on calcium phosphate columns. Arch Biochem Biophys 1956;65:132–55.
16. Bernardi G. Chromatography of nucleic acids on hydroxyapatite. Nature 1965;206: 779–83.
17. Khosravinia H, Ramesha KP. Influence of EDTA and magnesium on DNA extraction from blood samples and specificity of polymerase chain reaction. Afr J Biotechnol 2007;6:184–7.
18. Huggett JF, Novak T, Garson JA, et al. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes 2008;1:70.
19. Burger J, Hummel S, Hermann B, Henke W. DNA preservation: a microsatellite-DNA study on ancient skeletal remains. Electrophoresis 1999;20:1722–8.
20. Poly F, Chenu C, Simonet P, et al. Differences between linear chromosomal and supercoiled plasmid DNA in their mechanisms and extent of adsorption on clay minerals. Langmuir 2000;16:1233–8.
21. Pietramellara G, Franchi M, Gallori E, Nannipieri P. Effect of molecular characteristics of DNA on its adsorption and binding on homoionic montmorillonite and kaolinite. Biol Fert Soils 2001;33:402–9.
22. Goring CAI, Bartholomew WV. Adsorption of mononucleotides, nucleic acids, and nucleoproteins by clays. Soil Sci 1952;74:149–64.
23. Greaves MP, Wilson MJ. The adsorption of nucleic acids by montmorillonite. Soil Biol Biochem 1969;1:317–23.
24. Khanna M, Stotzky G. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl Environ Microbiol 1992;58:1930–9.
25. Wiechmann I, Grupe G. Detection of Yersinia pestis DNA in two early medieval skeletal finds from Aschheim (Upper Bavaria, 6th century A.D. Am J Phys Anthropol 2005;126:48–55.
26. Matheson CD, Vernon KK, Lahti A, et al. Molecular exploration of the first-century Tomb of the Shroud in Akeldama, Jerusalem. PLoS One 2009;4:e8319.
27. Miyauchi K, Ogawa M, Murata A, et al. Serum deoxyribonuclease I determined by a radioimmunoassay and an enzymatic assay in malignant diseases. Clin Chim Acta 1989;184:115–9.
28. Aitken ML, Burke W, McDonald G, et al. Recombinant human DNase inhalation in normal subjects and patients with cystic fibrosis. A phase 1 study. JAMA 1992;267:1947–51.
29. Brehm SP, Hoch SO, Hoch JA. DNA-binding proteins in human serum. Biochem Biophys Res Commun 1975;63:24–31.
30. Malonga H, Neault JF, Arakawa H, Tajmir-Riahi HA. DNA interaction with human serum albumin studied by affinity capillary electrophoresis and FTIR spectroscopy. DNA Cell Biol 2006;25:63–8.
31. Cai P, Huang Q, Li M, Liang W. Binding and degradation of DNA on montmorillonite coated by hydroxyl aluminum species. Colloids Surf B Biointerfaces 2008;62: 299–306.
32. Demanèche S, Jocteur-Monrozier L, Quiquampoix H, Simonet P. Evaluation of biological and physical protection against nuclease degradation of clay-bound plasmid DNA. Appl Environ Microbiol 2001;67:293–9.
33. Young G, Turner S, Davies JK, et al. Bacterial DNA persists for extended periods after cell death. J Endod 2007;33:1417–20.
34. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science 2002;295:1487.
35. Martins M, Uppuluri P, Thomas DP, et al. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 2010;169:323–31.
36. Siqueira JF Jr, Rôçac IN. Polymerase chain reaction-based analysis of microorganisms associated with failed endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2004;97:85–94.
37. Gomes BP, Pinheiro ET, Sousa EL, et al. Enterococcus faecalis in dental root canals detected by culture and by polymerase chain reaction analysis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2006;102:247–53.
38. Sundqvist G, Figdor D, Persson S, Sjögren U. Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1998;85:86–93.
39. Molander A, Reit C, Dahlén G, Kvist T. Microbiological status of root-filled teeth with apical periodontitis. Int EndodJ 1998;31:1–7.
40. Sundqvist G, Figdor D. Life as an endodontic pathogen: ecological differences between the untreated and root-filled root canal. Endod Topics 2003;6:3–28.
41. Nair PN. Abusing technology? Culture-difficult microbes and microbial remnants. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2007;104:569–70.
42. Siqueira JF Jr. On the issue of uncultivated bacteria and dead cell detection by molecular methods: reply to Dr. Nair’s commentary. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2008;105:5–8. author reply 8–10.
43. Herrmann BHS. Introduction. In: Herrmann B, Hummel S, eds. Ancient DNA. New York: Springer Verlag; 1994:1–12.
From the *Department of Odontology/Endodontics, Faculty of Medicine, Umeå University, Umea, Sweden; and †Department of Microbiology, Monash University, Melbourne, Victoria, Australia.
Supported by the County of Västerbotten, Public Dental Health and the Australian Dental Research Foundation Inc.
Address requests for reprints to Dr Malin Brundin, Department of Odontology/Endodontics, Faculty of Medicine, Umeå University, Umeå, 90187 Sweden. E-mail address: [email protected]
0099-2399/$ - see front matter
Copyright © 2013 American Association of Endodontists.
http://dx.doi.org/10.1016/j.joen.2012.09.013